Copper »
PDB 6l8s-6pvy »
6l9s »
Copper in PDB 6l9s: Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii
Protein crystallography data
The structure of Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii, PDB code: 6l9s
was solved by
C.Wang,
C.Y.Zhang,
Z.Z.Min,
X.L.Xu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.08 / 2.00 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 30.989, 57.420, 65.040, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.7 / 21.9 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii
(pdb code 6l9s). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii, PDB code: 6l9s:
In total only one binding site of Copper was determined in the Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii, PDB code: 6l9s:
Copper binding site 1 out of 1 in 6l9s
Go back to
Copper binding site 1 out
of 1 in the Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii
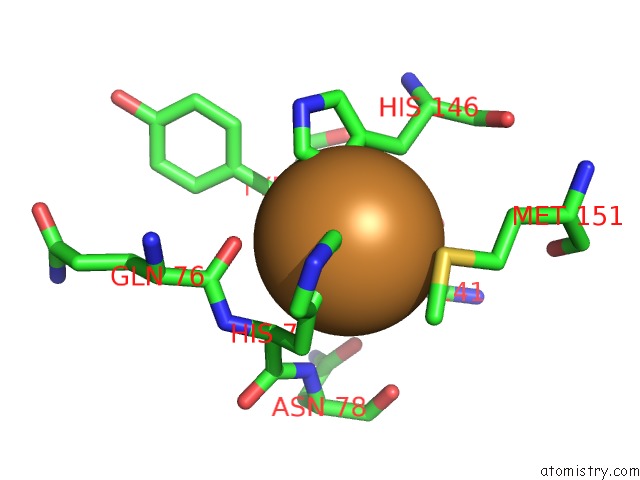
Mono view
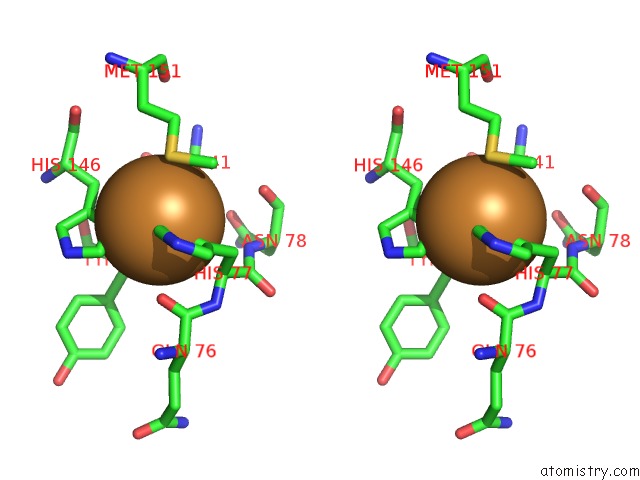
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of Na-Dithionite Reduced Auracyanin From Photosynthetic Bacterium Roseiflexus Castenholzii within 5.0Å range:
|
Reference:
C.Wang,
Y.Y.Xin,
Z.Z.Min,
J.Qi,
C.Y.Zhang,
X.L.Xu.
Structural Basis Underlying the Electron Transfer Features of A Blue Copper Protein Auracyanin From the Photosynthetic Bacterium Roseiflexus Castenholzii. Photosyn. Res. 2020.
ISSN: ISSN 1573-5079
PubMed: 31933173
DOI: 10.1007/S11120-020-00709-Y
Page generated: Mon Jul 14 06:32:03 2025
ISSN: ISSN 1573-5079
PubMed: 31933173
DOI: 10.1007/S11120-020-00709-Y
Last articles
Hg in 1U19Hg in 1UGF
Hg in 1TLF
Hg in 1UGE
Hg in 1UGC
Hg in 1SMS
Hg in 1T83
Hg in 1RWA
Hg in 1T3S
Hg in 1S1F