Copper »
PDB 9csu-9j9q »
9hdg »
Copper in PDB 9hdg: The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus
Protein crystallography data
The structure of The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus, PDB code: 9hdg
was solved by
K.Hall,
S.E.Roennekleiv,
A.Gautieri,
R.Skaali,
L.Rieder,
A.N.Englund,
E.Landsem,
T.Emrich-Mills,
I.Ayuso-Fernandez,
O.Golten,
A.R.Kjendseth,
M.Soerlie,
V.G.H.Eijsink,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.46 / 1.26 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.332, 53.655, 70.929, 90, 90, 90 |
| R / Rfree (%) | 16.5 / 19.6 |
Copper Binding Sites:
The binding sites of Copper atom in the The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus
(pdb code 9hdg). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus, PDB code: 9hdg:
In total only one binding site of Copper was determined in the The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus, PDB code: 9hdg:
Copper binding site 1 out of 1 in 9hdg
Go back to
Copper binding site 1 out
of 1 in the The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus
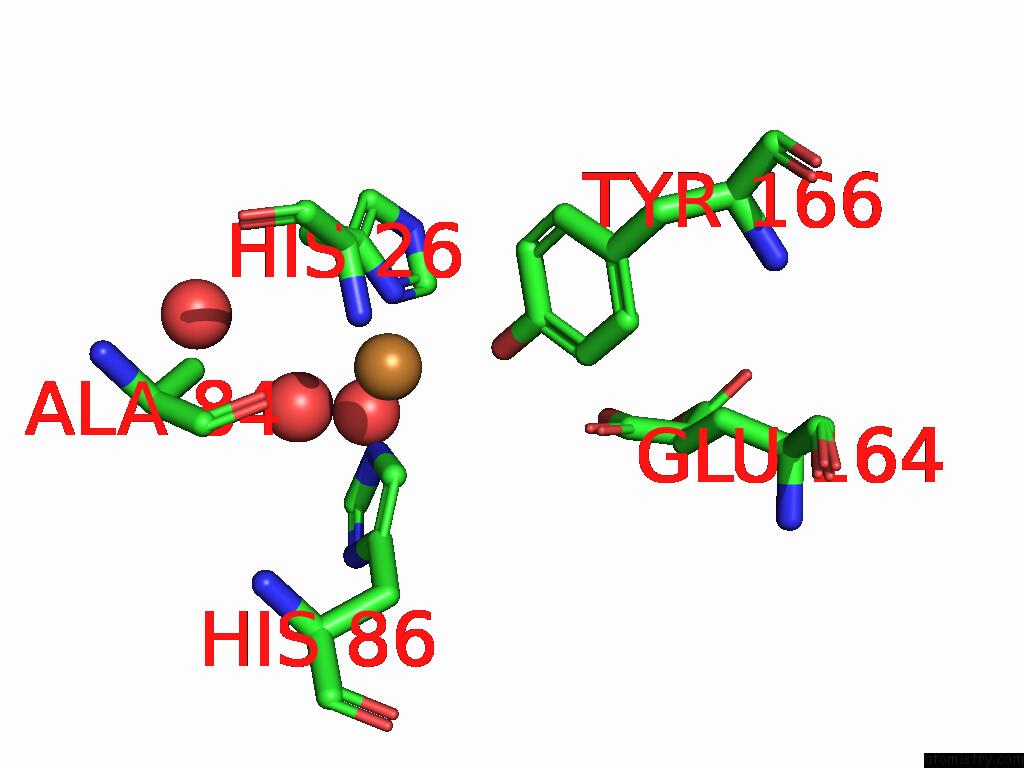
Mono view
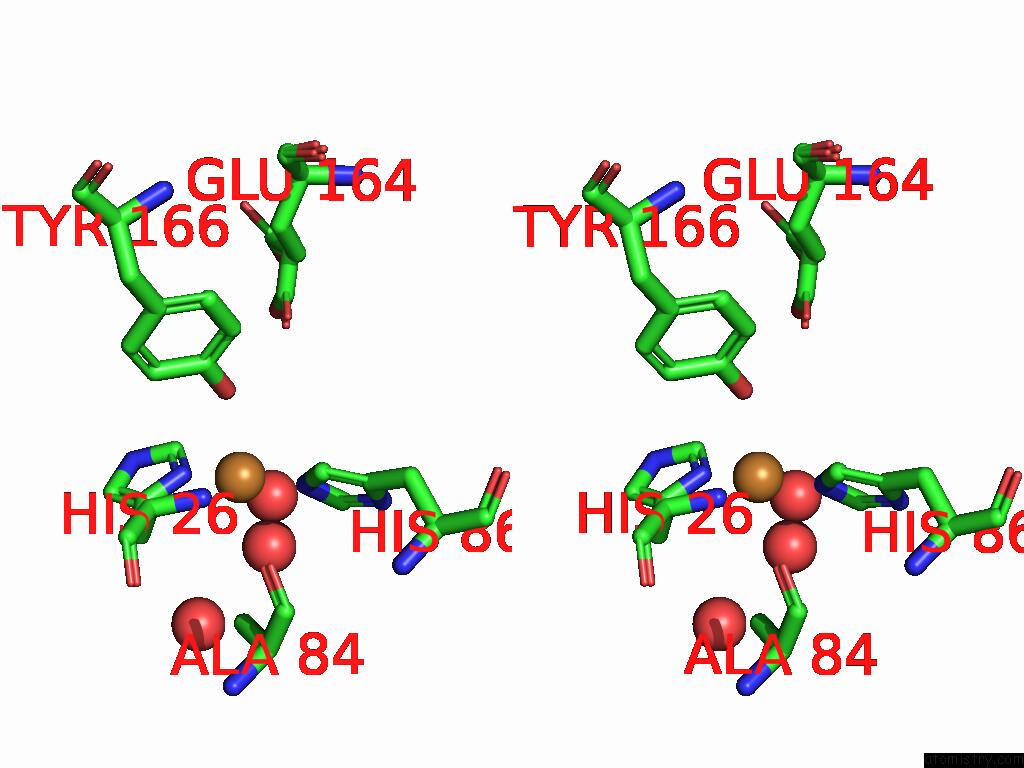
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of The Structure of the Catalytic Domain of AFAA11B From the Filamentous Fungus Aspergillus Fumigatus within 5.0Å range:
|
Reference:
K.R.Hall,
S.Elisa Ronnekleiv,
A.Gautieri,
H.Lilleas,
R.Skaali,
L.Rieder,
A.Nikoline Englund,
E.Landsem,
T.Z.Emrich-Mills,
I.Ayuso-Fernandez,
A.Kjendseth Rohr,
M.Sorlie,
V.G.H.Eijsink.
Structure-Function Analysis of An Understudied Type of Lpmo with Unique Redox Properties and Substrate Specificity. Acs Catalysis V. 15 10601 2025.
ISSN: ESSN 2155-5435
PubMed: 40568223
DOI: 10.1021/ACSCATAL.5C03003
Page generated: Mon Jul 14 09:57:14 2025
ISSN: ESSN 2155-5435
PubMed: 40568223
DOI: 10.1021/ACSCATAL.5C03003
Last articles
Hg in 1R7UHg in 1R7T
Hg in 1QZ4
Hg in 1PM2
Hg in 1R76
Hg in 1QD9
Hg in 1R0G
Hg in 1QML
Hg in 1QAI
Hg in 1PJ1