Copper »
PDB 9csu-9j9q »
9ha0 »
Copper in PDB 9ha0: Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs
Protein crystallography data
The structure of Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs, PDB code: 9ha0
was solved by
A.M.W.H.Thunnissen,
R.Jiang,
F.Casilli,
F.Aalbers,
G.Roelfes,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.43 / 1.75 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 35.395, 54.313, 145.734, 90, 90, 90 |
| R / Rfree (%) | 19.8 / 23.5 |
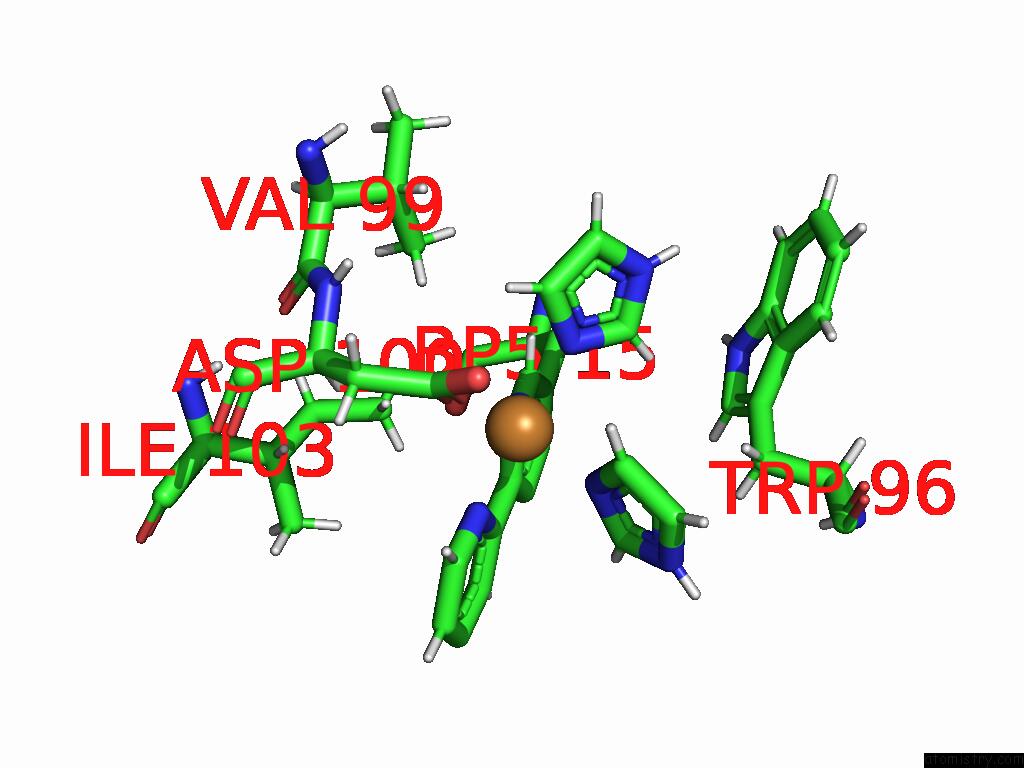
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs
(pdb code 9ha0). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs, PDB code: 9ha0:
In total only one binding site of Copper was determined in the Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs, PDB code: 9ha0:
Copper binding site 1 out of 1 in 9ha0
Go back to
Copper binding site 1 out
of 1 in the Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs
Mono view
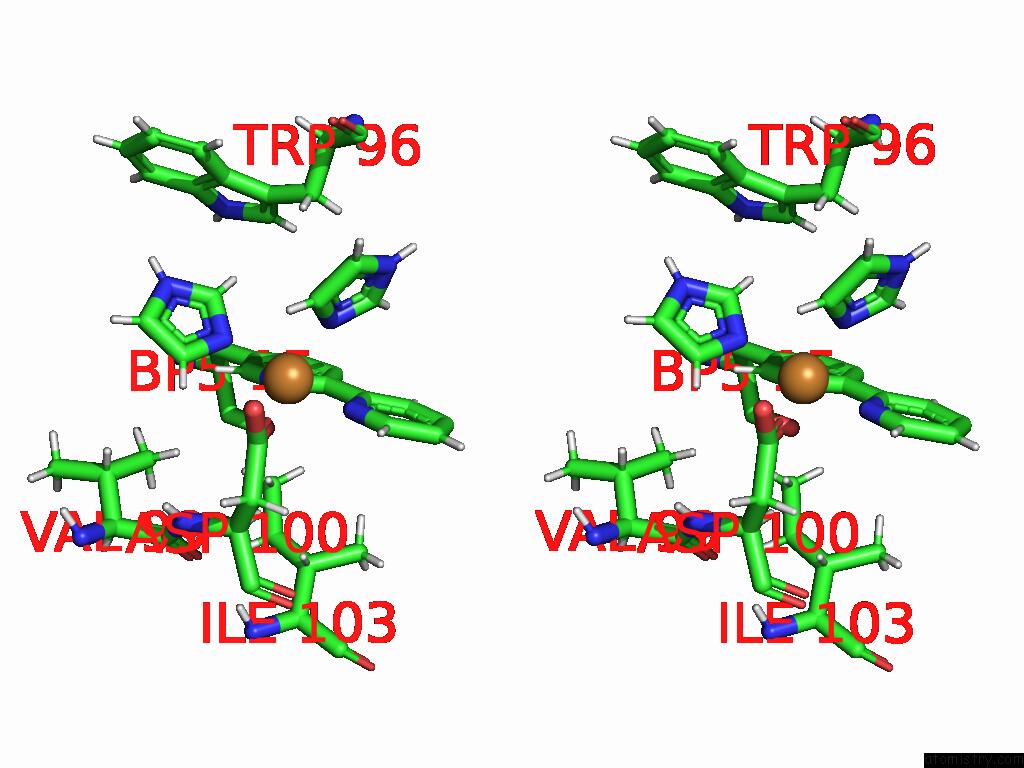
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of Cu(II)-Bound LMRR_V15BPY Variant Bvs within 5.0Å range:
|
Reference:
R.Jiang,
F.Casilli,
A.W.H.Thunnissen,
G.Roelfes.
An Artificial Copper-Michaelase Featuring A Genetically Encoded Bipyridine Ligand For Asymmetric Additions to Nitroalkenes. Angew.Chem.Int.Ed.Engl. 23182 2025.
ISSN: ESSN 1521-3773
PubMed: 39945539
DOI: 10.1002/ANIE.202423182
Page generated: Mon Jul 14 09:57:12 2025
ISSN: ESSN 1521-3773
PubMed: 39945539
DOI: 10.1002/ANIE.202423182
Last articles
Hg in 1R76Hg in 1QD9
Hg in 1R0G
Hg in 1QML
Hg in 1QAI
Hg in 1PJ1
Hg in 1QBZ
Hg in 1PTK
Hg in 1PVM
Hg in 1PPO