Copper »
PDB 8uem-9cst »
8vhs »
Copper in PDB 8vhs: X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif
Protein crystallography data
The structure of X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif, PDB code: 8vhs
was solved by
S.Chakraborty,
S.Mitra,
D.Prakash,
P.Prasad,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 26.52 / 1.36 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 26.41, 45.47, 97.95, 90, 90, 90 |
| R / Rfree (%) | 19.6 / 22.5 |
Copper Binding Sites:
The binding sites of Copper atom in the X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif
(pdb code 8vhs). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif, PDB code: 8vhs:
In total only one binding site of Copper was determined in the X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif, PDB code: 8vhs:
Copper binding site 1 out of 1 in 8vhs
Go back to
Copper binding site 1 out
of 1 in the X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif
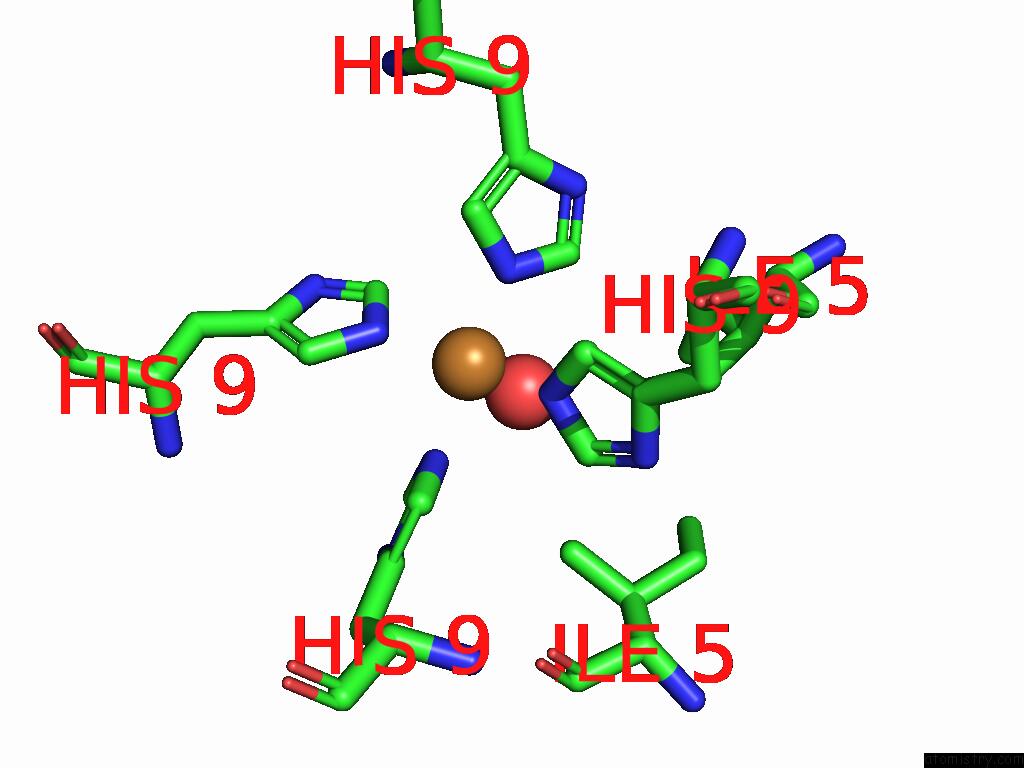
Mono view
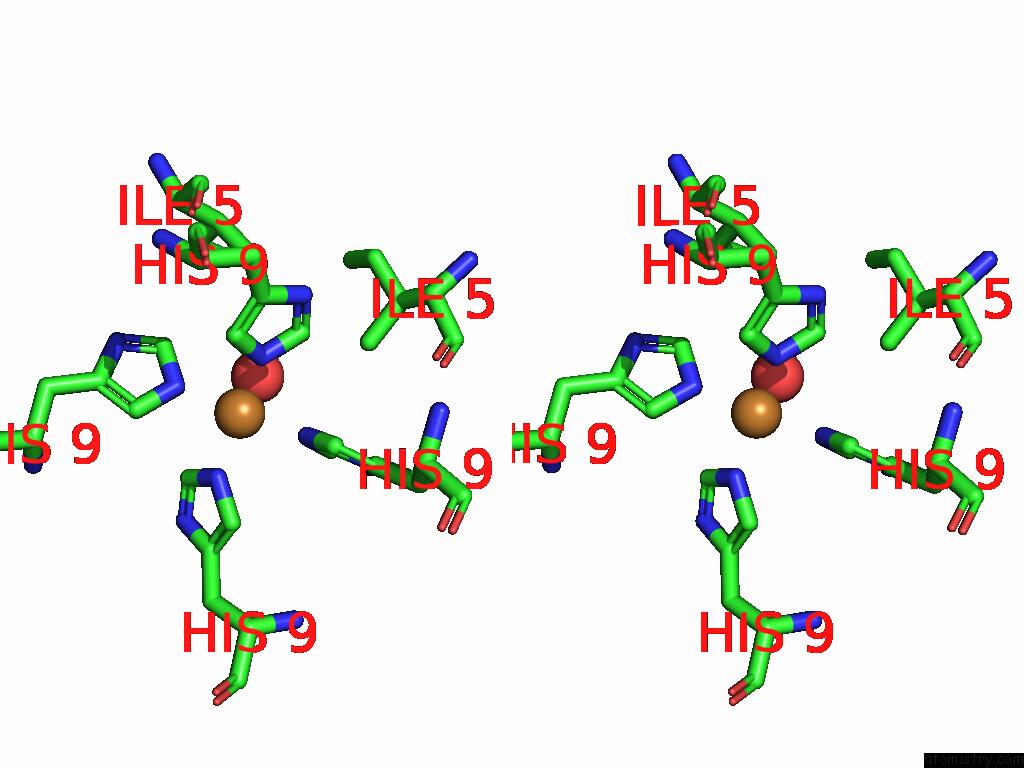
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of X-Ray Structure of A De Novo Designed Self Assembled Peptide Tetramer Featuring A Cu(His)4(H2O) Coordination Motif within 5.0Å range:
|
Reference:
D.Prakash,
S.Mitra,
S.Sony,
M.Murphy,
B.Andi,
L.Ashley,
P.Prasad,
S.Chakraborty.
Controlling Outer-Sphere Solvent Reorganization Energy to Turn on or Off the Function of Artificial Metalloenzymes. Nat Commun V. 16 3048 2025.
ISSN: ESSN 2041-1723
PubMed: 40155633
DOI: 10.1038/S41467-025-57904-5
Page generated: Mon Jul 14 09:37:50 2025
ISSN: ESSN 2041-1723
PubMed: 40155633
DOI: 10.1038/S41467-025-57904-5
Last articles
I in 1XBUI in 1XB7
I in 1WY4
I in 1WQ3
I in 1WCB
I in 1WCV
I in 1WC7
I in 1VKC
I in 1W51
I in 1W50