Copper »
PDB 7s1f-7xmb »
7t4p »
Copper in PDB 7t4p: Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
Enzymatic activity of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
All present enzymatic activity of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution:
1.14.13.25; 1.14.18.3;
1.14.13.25; 1.14.18.3;
Copper Binding Sites:
The binding sites of Copper atom in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
(pdb code 7t4p). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 9 binding sites of Copper where determined in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution, PDB code: 7t4p:
Jump to Copper binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
In total 9 binding sites of Copper where determined in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution, PDB code: 7t4p:
Jump to Copper binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9;
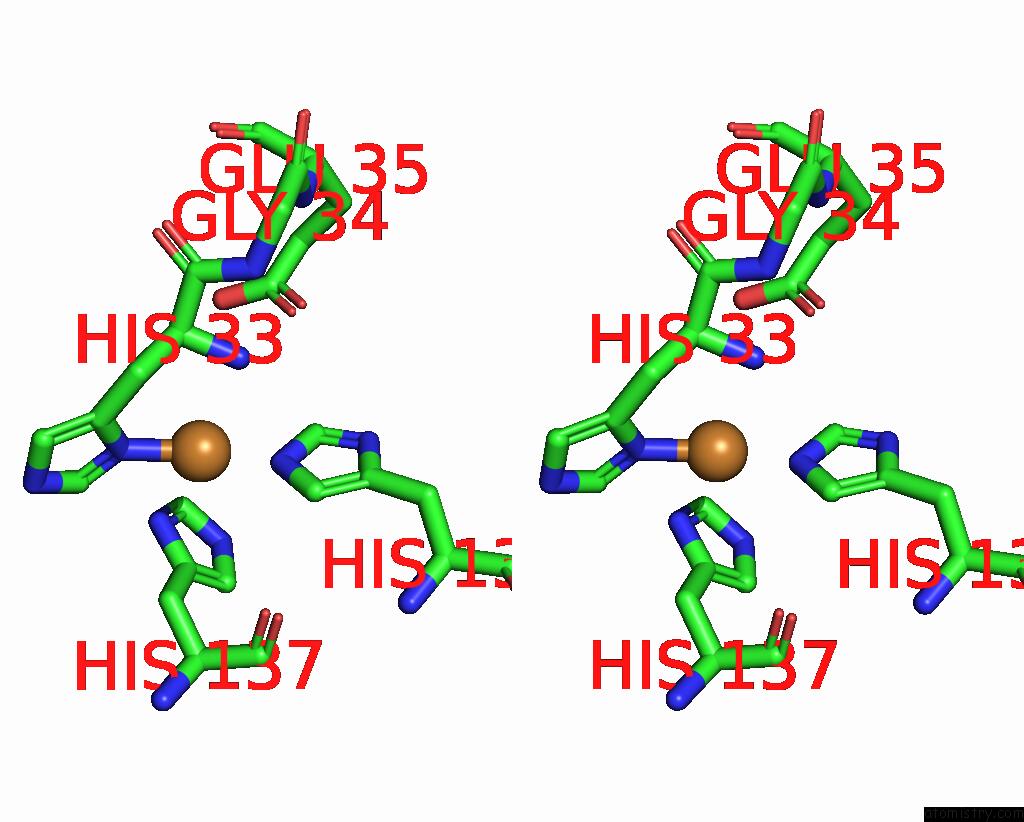
Copper binding site 1 out of 9 in 7t4p
Go back to
Copper binding site 1 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
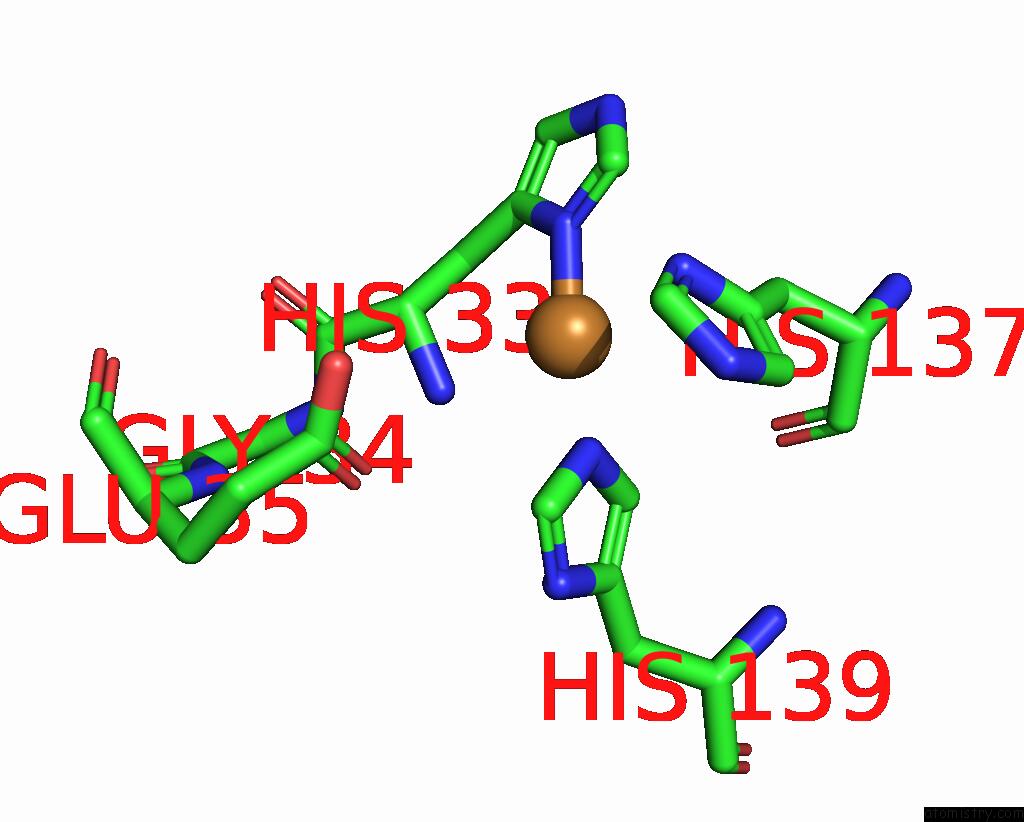
Mono view
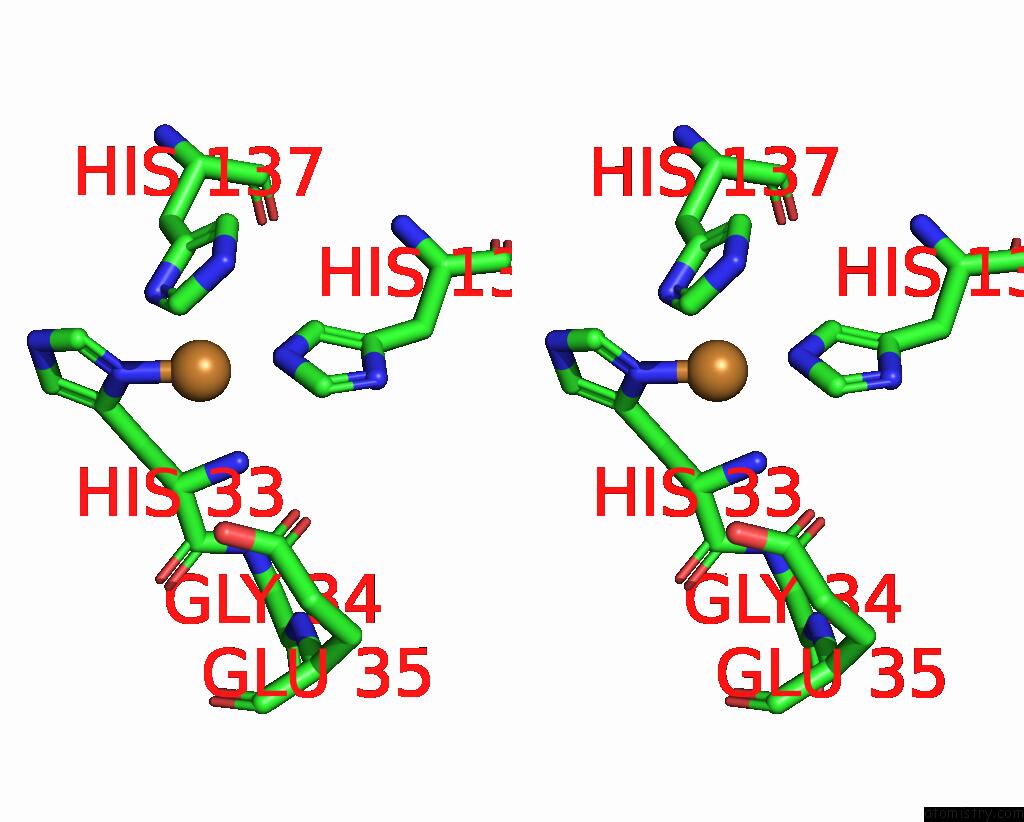
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 2 out of 9 in 7t4p
Go back to
Copper binding site 2 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 3 out of 9 in 7t4p
Go back to
Copper binding site 3 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
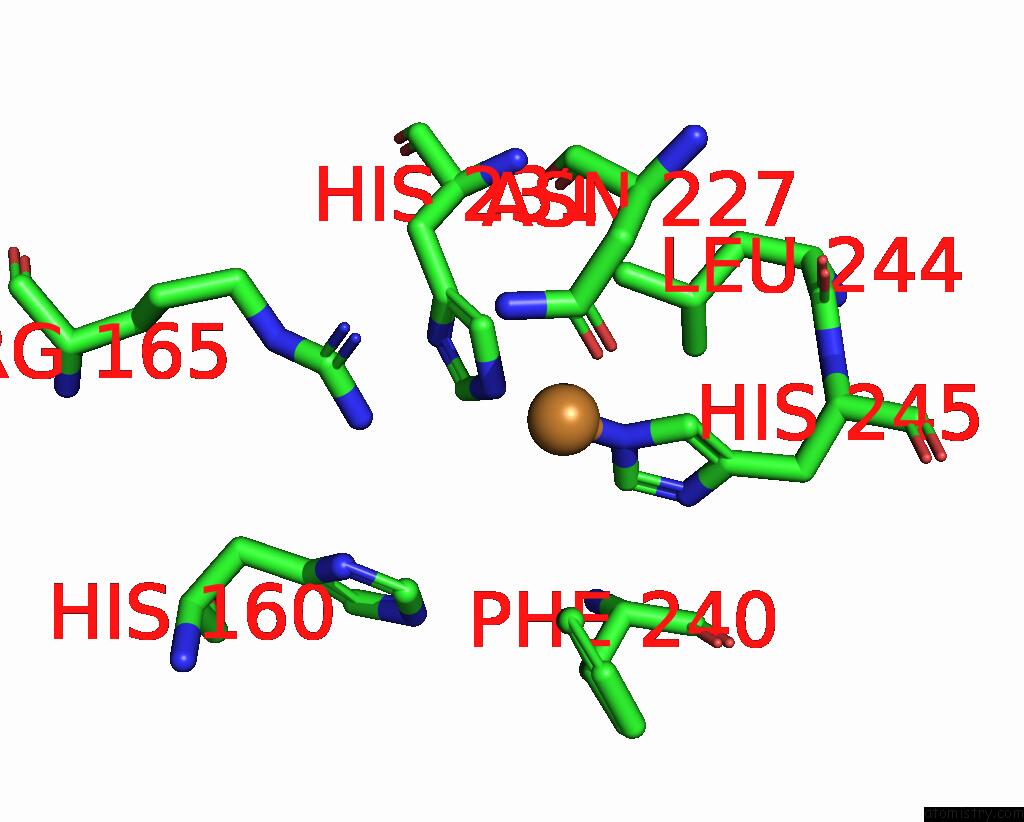
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 3 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 4 out of 9 in 7t4p
Go back to
Copper binding site 4 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
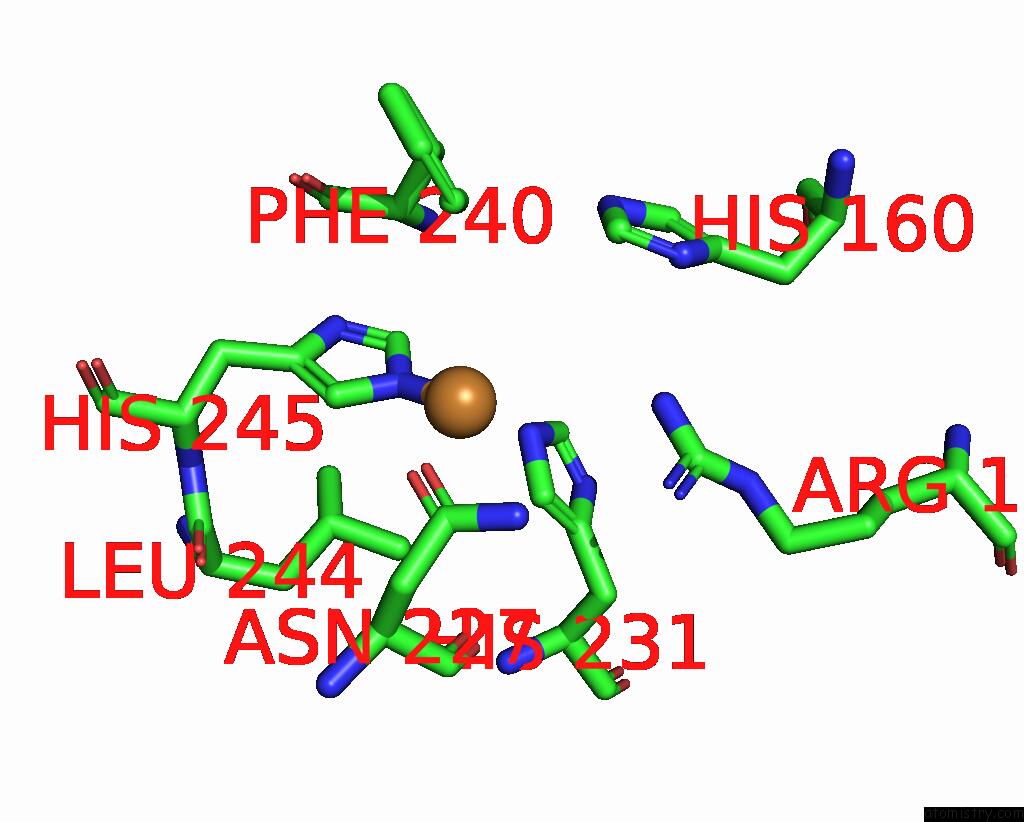
Mono view
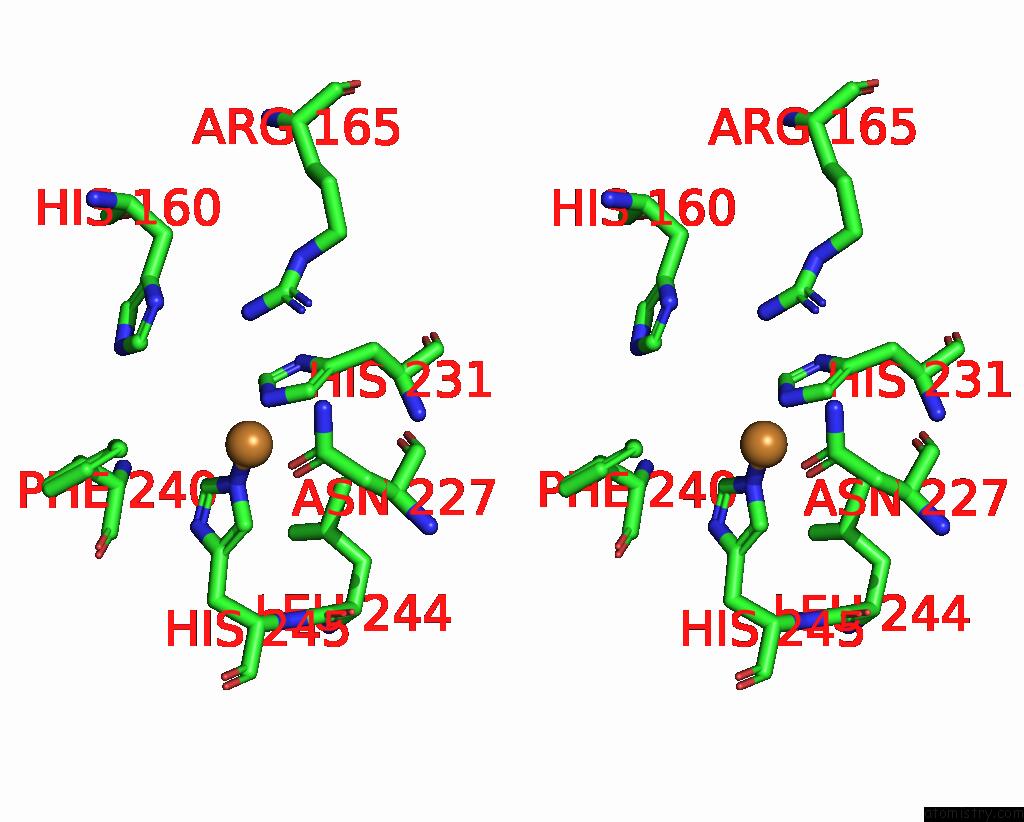
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 4 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 5 out of 9 in 7t4p
Go back to
Copper binding site 5 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
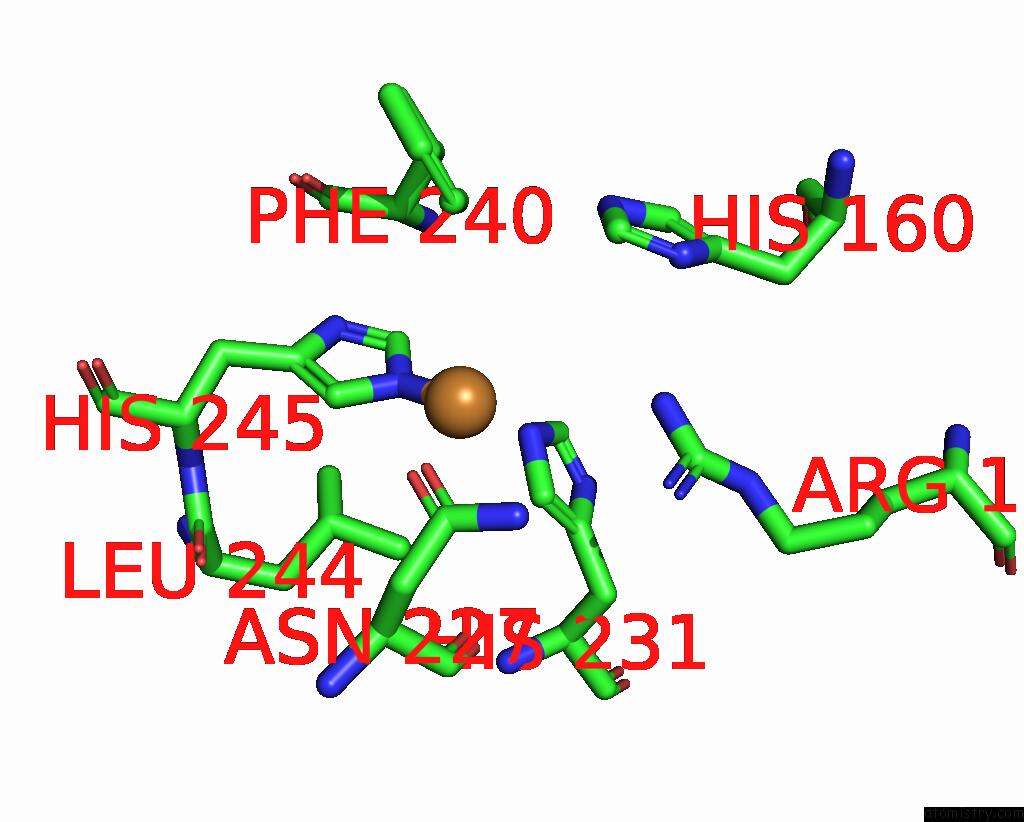
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 5 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
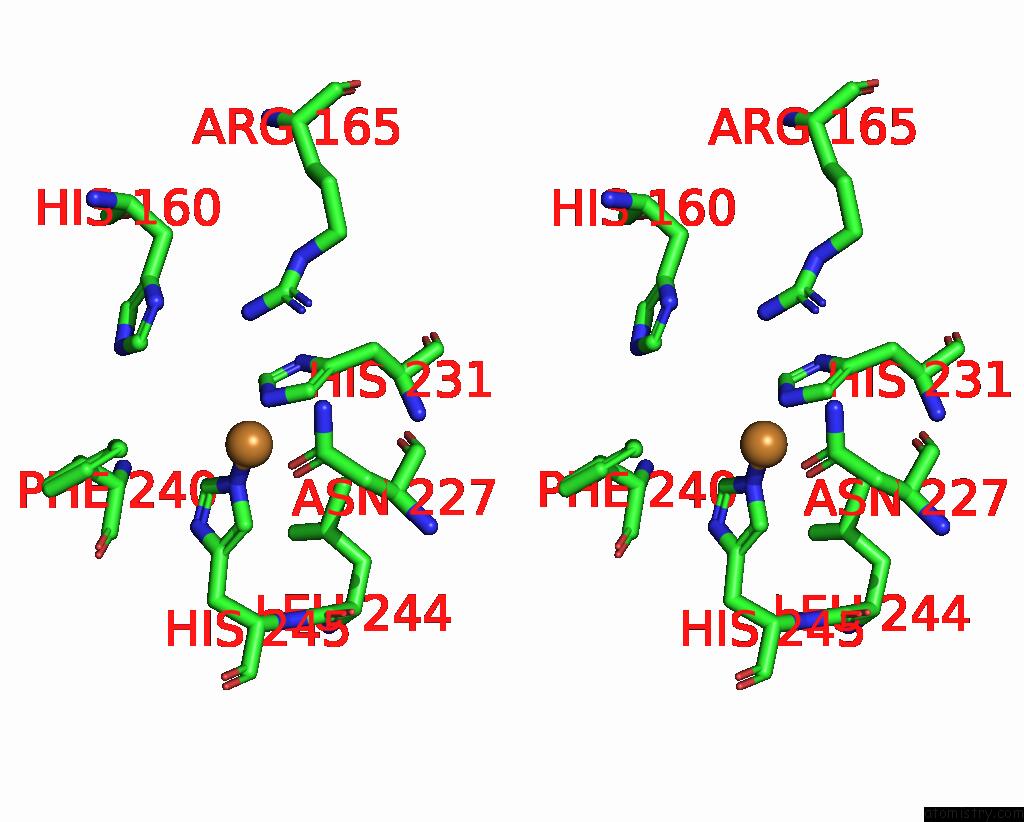
Copper binding site 6 out of 9 in 7t4p
Go back to
Copper binding site 6 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 6 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 7 out of 9 in 7t4p
Go back to
Copper binding site 7 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
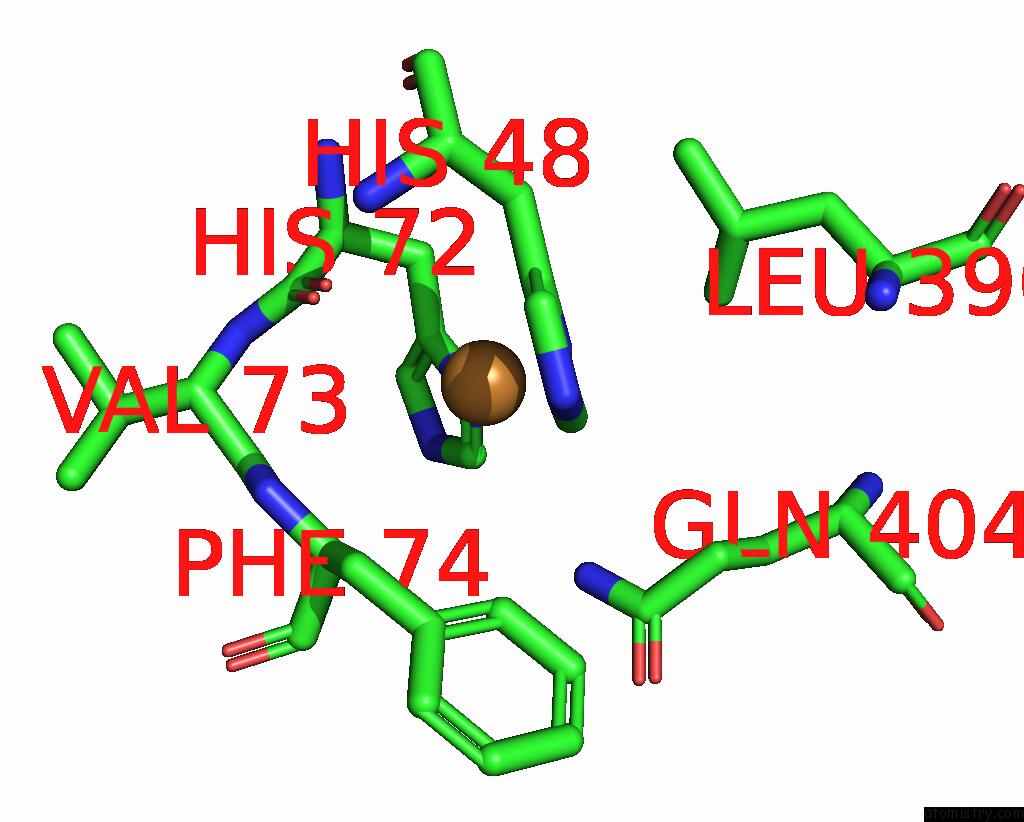
Mono view
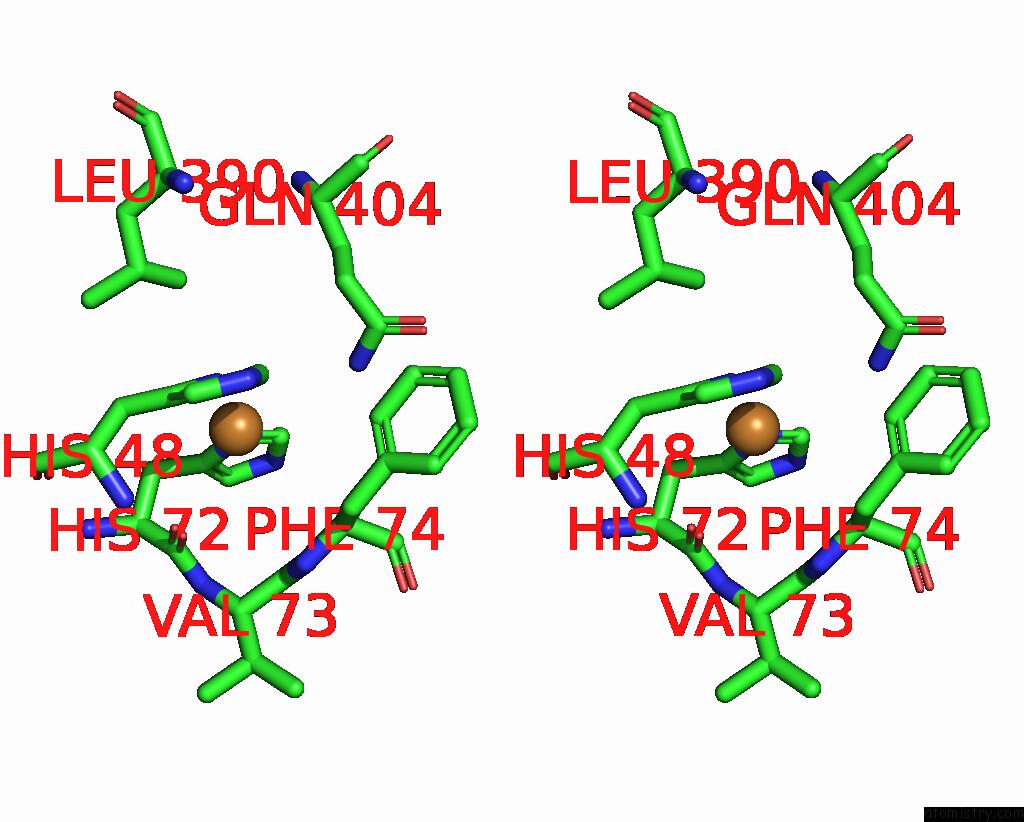
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 7 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 8 out of 9 in 7t4p
Go back to
Copper binding site 8 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
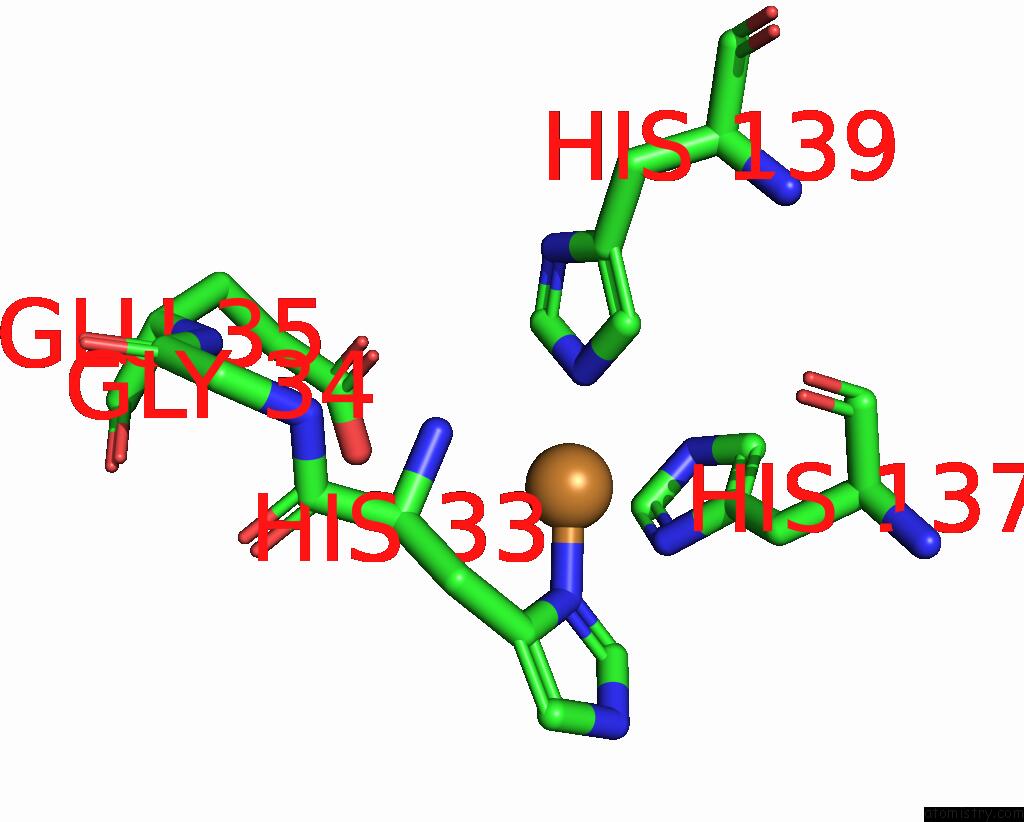
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 8 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Copper binding site 9 out of 9 in 7t4p
Go back to
Copper binding site 9 out
of 9 in the Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution
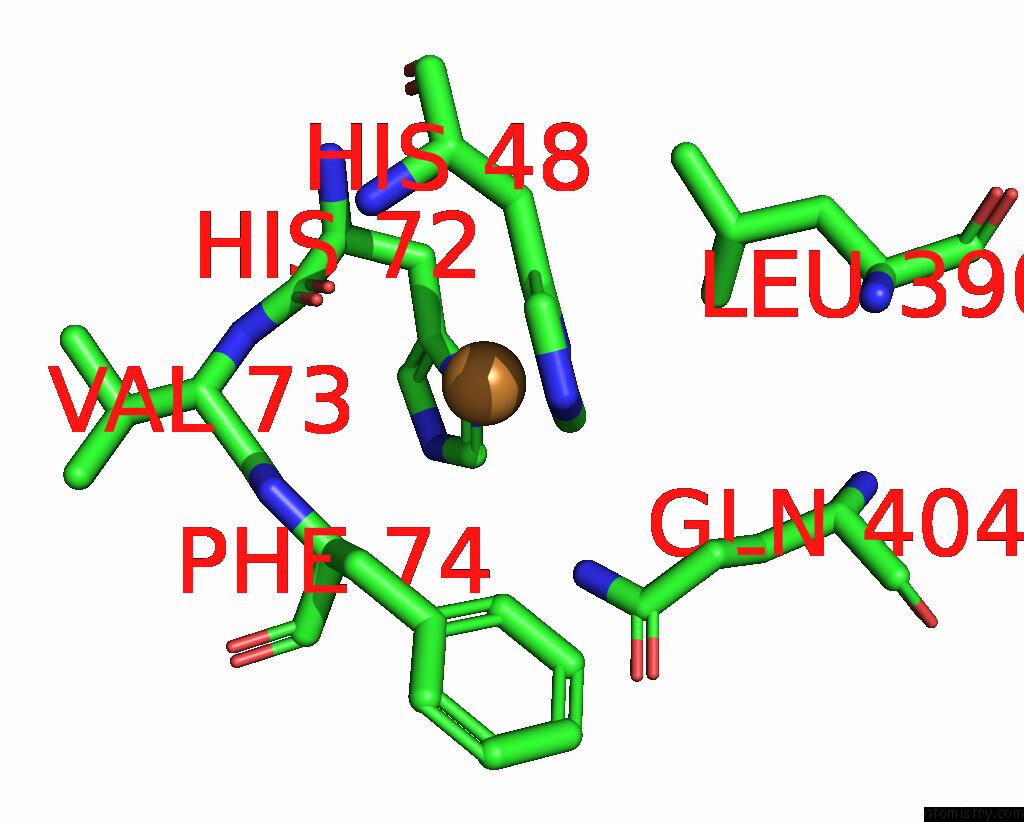
Mono view
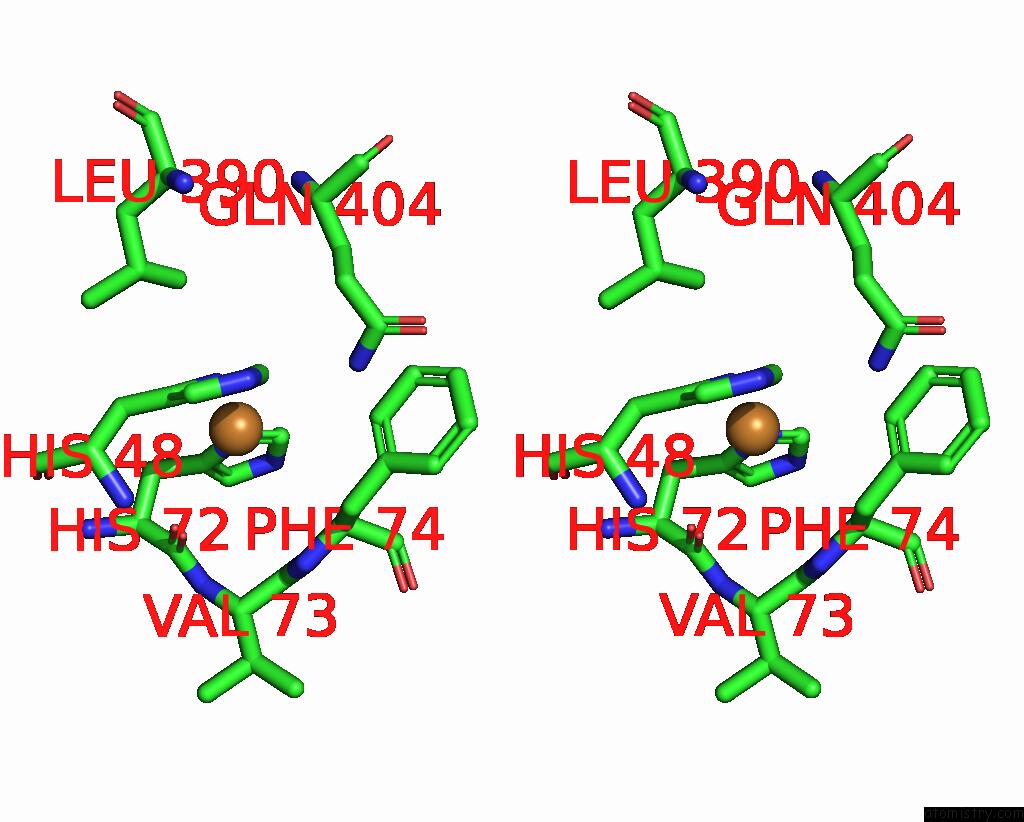
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 9 of Cryoem Structure of Methylococcus Capsulatus (Bath) Pmmo Treated with Potassium Cyanide and Copper in A Native Lipid Nanodisc at 3.62 Angstrom Resolution within 5.0Å range:
|
Reference:
C.W.Koo,
F.J.Tucci,
Y.He,
A.C.Rosenzweig.
Recovery of Particulate Methane Monooxygenase Structure and Activity in A Lipid Bilayer. Science V. 375 1287 2022.
ISSN: ESSN 1095-9203
PubMed: 35298269
DOI: 10.1126/SCIENCE.ABM3282
Page generated: Mon Jul 14 08:33:13 2025
ISSN: ESSN 1095-9203
PubMed: 35298269
DOI: 10.1126/SCIENCE.ABM3282
Last articles
I in 2CIWI in 2C3N
I in 2C6C
I in 2C3V
I in 2C47
I in 2BXN
I in 2BIV
I in 2B9X
I in 2BXL
I in 2BMK