Copper »
PDB 7pyl-7s1d »
7s1c »
Copper in PDB 7s1c: Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1)
Protein crystallography data
The structure of Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1), PDB code: 7s1c
was solved by
B.Heras,
M.J.Scanlon,
J.L.Martin,
P.Sharma,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.95 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.199, 63.701, 74.257, 90, 126.11, 90 |
| R / Rfree (%) | 16.7 / 20.9 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1)
(pdb code 7s1c). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1), PDB code: 7s1c:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1), PDB code: 7s1c:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 7s1c
Go back to
Copper binding site 1 out
of 2 in the Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1)

Mono view
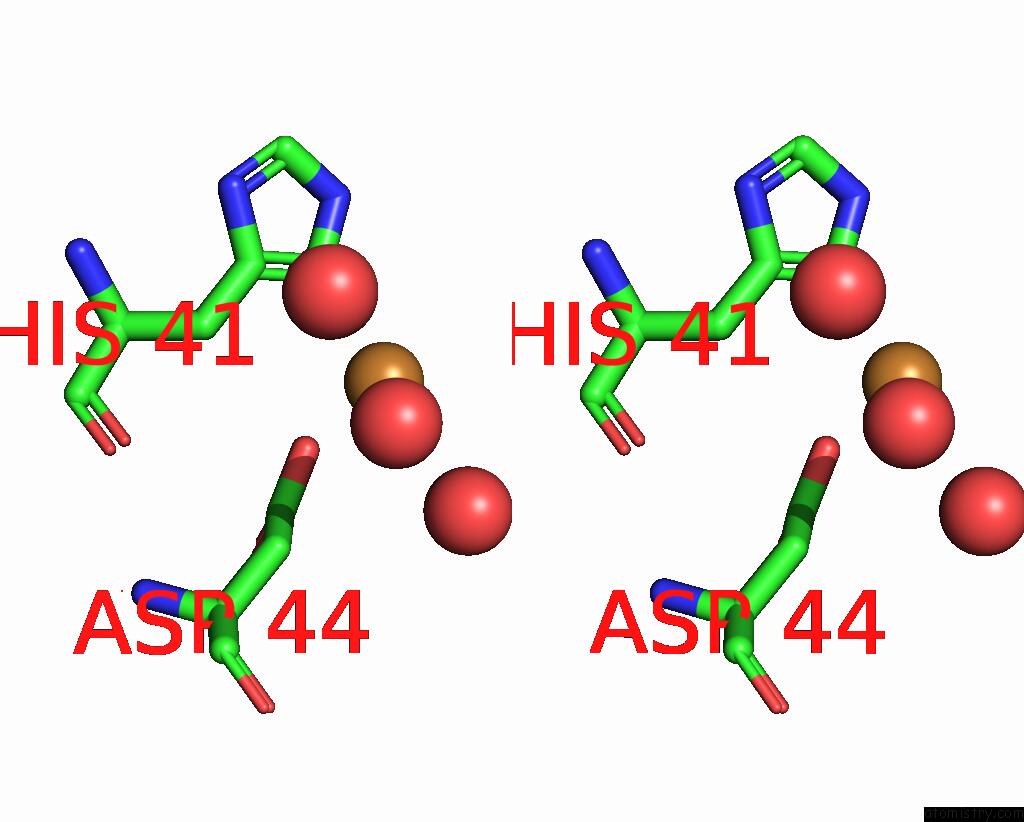
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1) within 5.0Å range:
|
Copper binding site 2 out of 2 in 7s1c
Go back to
Copper binding site 2 out
of 2 in the Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1)
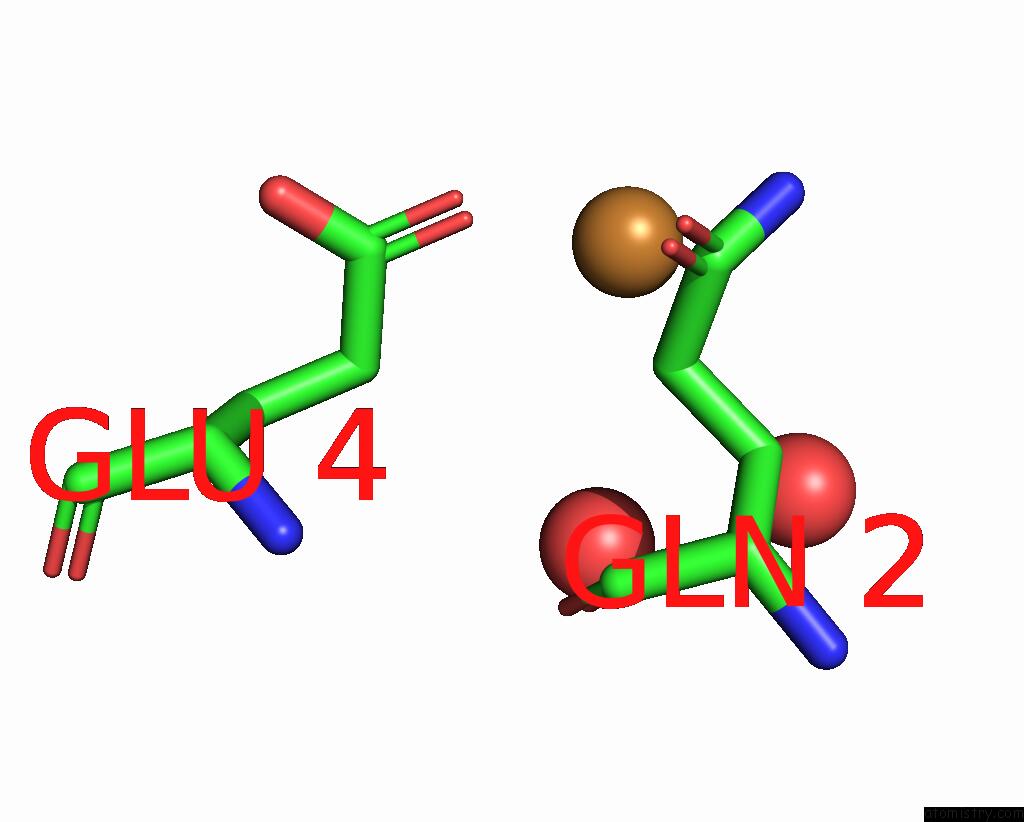
Mono view
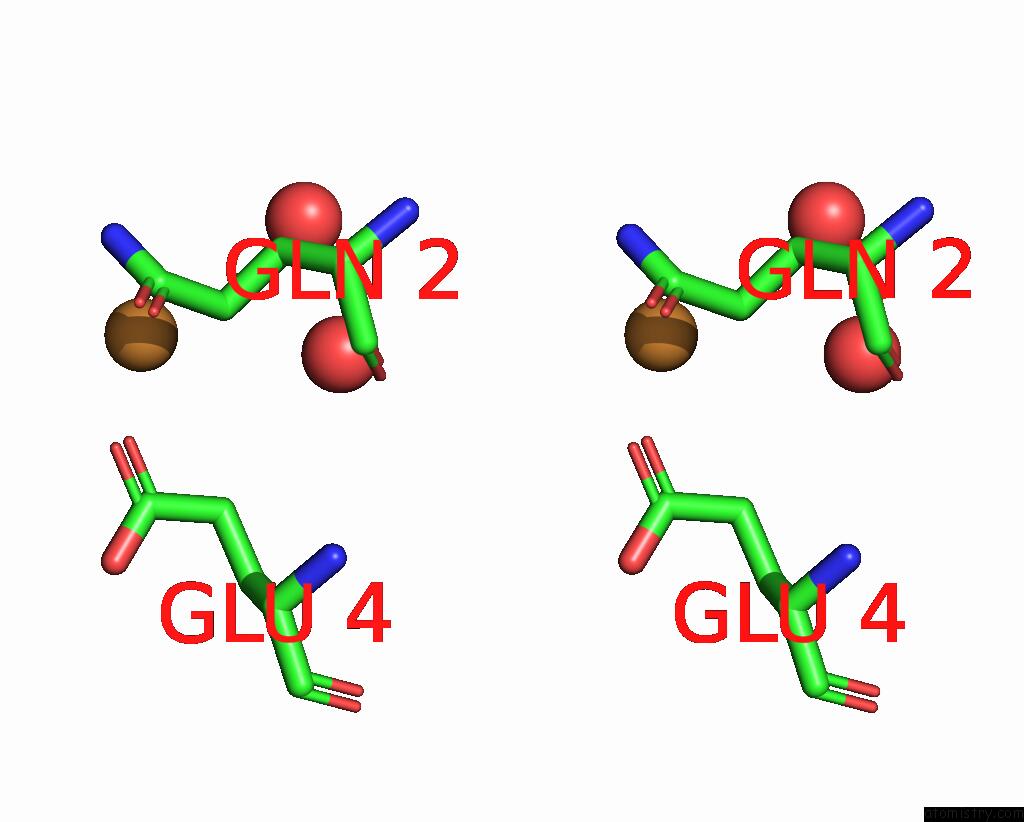
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of E.Coli Dsba in Complex with Compound Mips-0001897 (Compound 1) within 5.0Å range:
|
Reference:
C.D.Bradley,
R.L.Whitehouse,
K.Rimmer,
M.Williams,
B.Heras,
S.Caria,
O.Ilyichova,
M.Vazirani,
B.Mohanty,
J.Harper,
M.J.Scanlon,
J.S.Simpson.
Fluoromethylketone-Fragment Conjugates Designed As Covalent Modifiers of Ecdsba Are Atypical Substrates Chemrxiv 2022.
ISSN: ISSN 2573-2293
DOI: 10.26434/CHEMRXIV-2022-262LH
Page generated: Mon Jul 14 08:29:23 2025
ISSN: ISSN 2573-2293
DOI: 10.26434/CHEMRXIV-2022-262LH
Last articles
K in 5WIEK in 5WNN
K in 5WK9
K in 5WK7
K in 5WJN
K in 5WGM
K in 5WJM
K in 5WJ8
K in 5WJ1
K in 5WDL