Copper »
PDB 5onx-5wbc »
5tki »
Copper in PDB 5tki: Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement
Protein crystallography data
The structure of Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement, PDB code: 5tki
was solved by
W.B.O'dell,
F.Meilleur,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | N/A / 1.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 68.120, 42.230, 70.290, 90.00, 98.33, 90.00 |
| R / Rfree (%) | 21.6 / 25.3 |
Copper Binding Sites:
The binding sites of Copper atom in the Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement
(pdb code 5tki). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement, PDB code: 5tki:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement, PDB code: 5tki:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 5tki
Go back to
Copper binding site 1 out
of 2 in the Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement
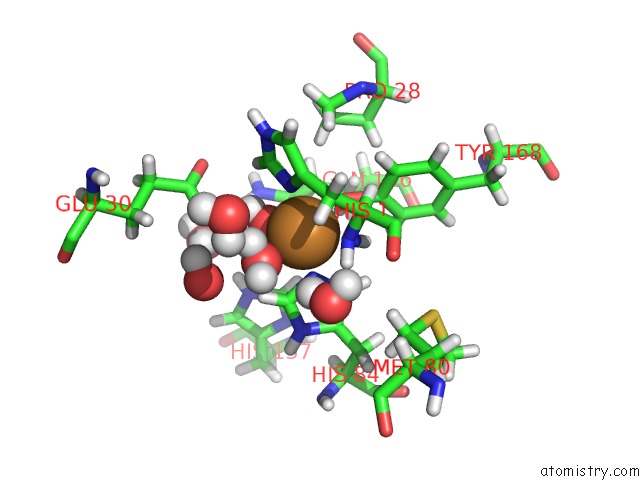
Mono view
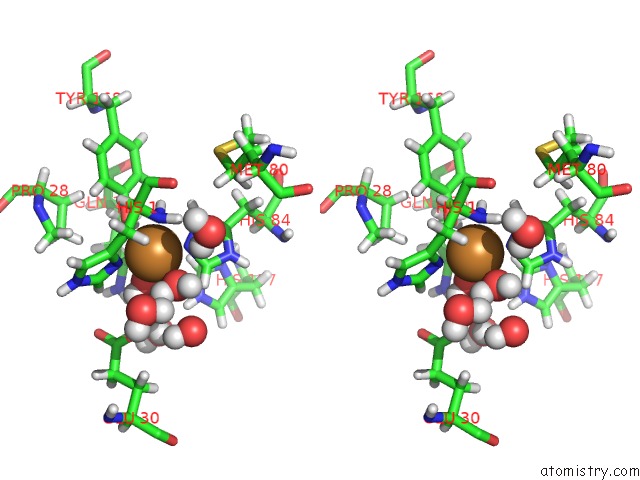
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement within 5.0Å range:
|
Copper binding site 2 out of 2 in 5tki
Go back to
Copper binding site 2 out
of 2 in the Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement
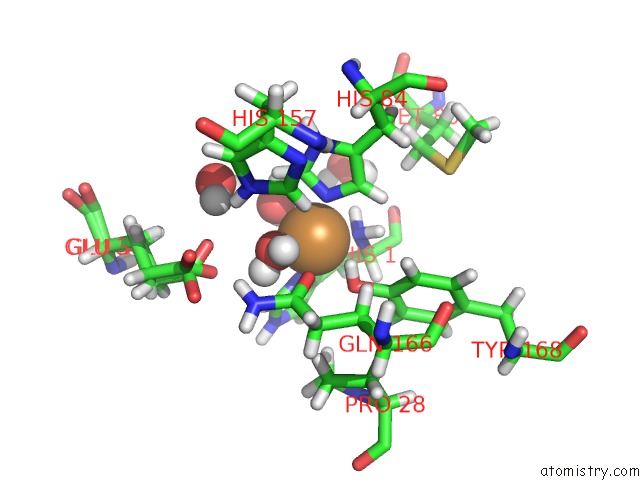
Mono view
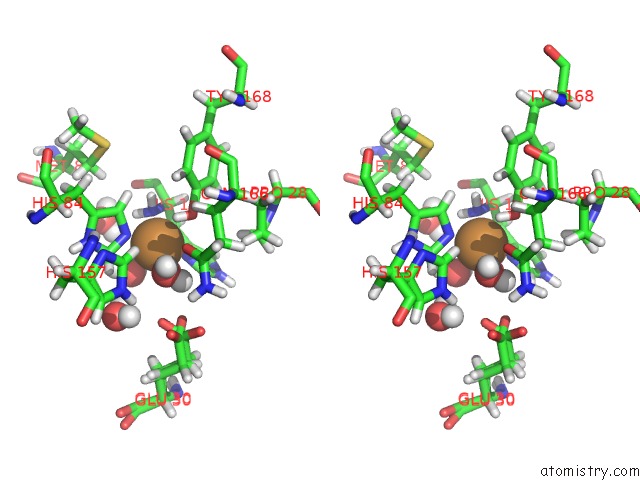
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Neurospora Crassa Polysaccharide Monooxygenase 2 Resting State Joint X-Ray/Neutron Refinement within 5.0Å range:
|
Reference:
W.B.O'dell,
P.K.Agarwal,
F.Meilleur.
Oxygen Activation at the Active Site of A Fungal Lytic Polysaccharide Monooxygenase. Angew. Chem. Int. Ed. Engl. V. 56 767 2017.
ISSN: ESSN 1521-3773
PubMed: 28004877
DOI: 10.1002/ANIE.201610502
Page generated: Mon Jul 14 05:25:50 2025
ISSN: ESSN 1521-3773
PubMed: 28004877
DOI: 10.1002/ANIE.201610502
Last articles
I in 5F1TI in 5FOR
I in 5F1W
I in 5EOJ
I in 5EUZ
I in 5ENM
I in 5ENK
I in 5EEK
I in 5E9O
I in 5EMS