Copper »
PDB 4tm7-4yst »
4yss »
Copper in PDB 4yss: Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy
Protein crystallography data
The structure of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy, PDB code: 4yss
was solved by
Y.Fukuda,
K.M.Tse,
M.Suzuki,
K.Diedrichs,
K.Hirata,
T.Nakane,
M.Sugahara,
E.Nango,
K.Tono,
Y.Joti,
T.Kameshima,
C.Song,
T.Hatsui,
M.Yabashi,
O.Nureki,
H.Matsumura,
T.Inoue,
S.Iwata,
E.Mizohata,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 33.23 / 1.50 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 115.098, 115.098, 84.305, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 14.8 / 19.2 |
Copper Binding Sites:
The binding sites of Copper atom in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy
(pdb code 4yss). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 7 binding sites of Copper where determined in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy, PDB code: 4yss:
Jump to Copper binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Copper where determined in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy, PDB code: 4yss:
Jump to Copper binding site number: 1; 2; 3; 4; 5; 6; 7;
Copper binding site 1 out of 7 in 4yss
Go back to
Copper binding site 1 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
Copper binding site 2 out of 7 in 4yss
Go back to
Copper binding site 2 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
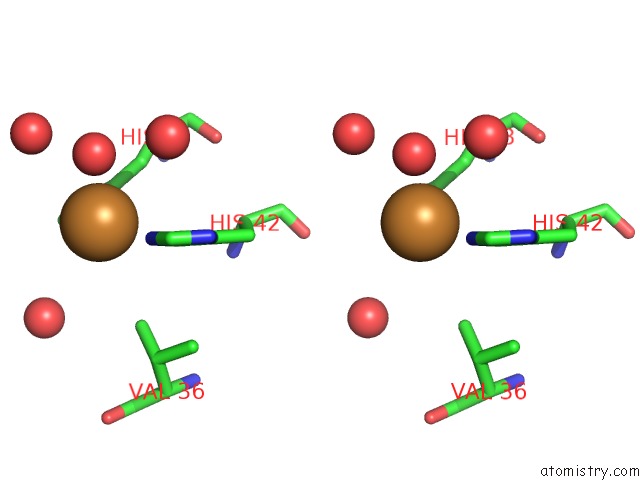
Copper binding site 3 out of 7 in 4yss
Go back to
Copper binding site 3 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 3 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
Copper binding site 4 out of 7 in 4yss
Go back to
Copper binding site 4 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 4 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
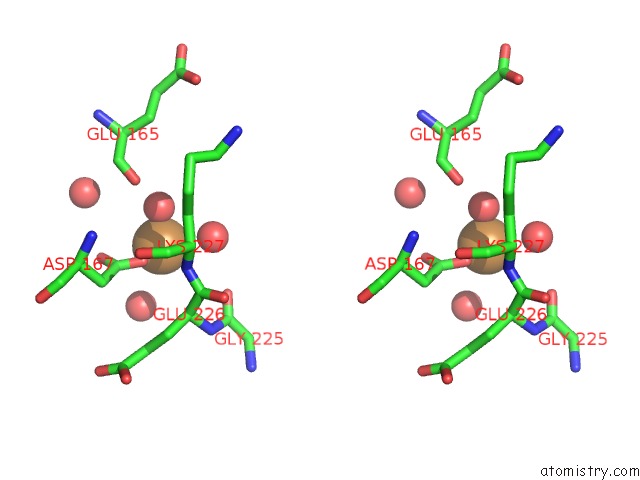
Copper binding site 5 out of 7 in 4yss
Go back to
Copper binding site 5 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 5 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
Copper binding site 6 out of 7 in 4yss
Go back to
Copper binding site 6 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 6 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
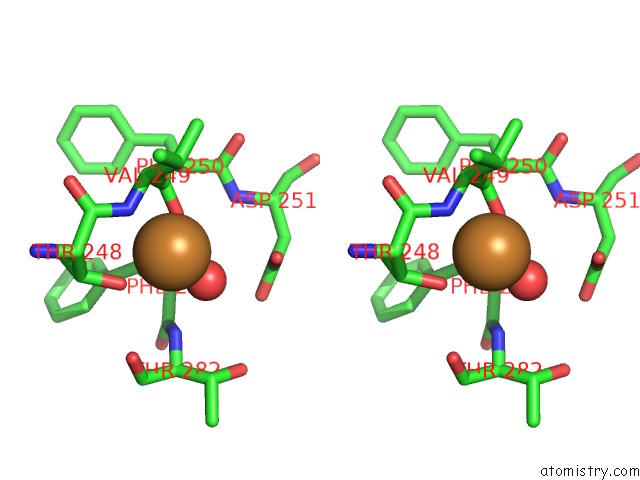
Copper binding site 7 out of 7 in 4yss
Go back to
Copper binding site 7 out
of 7 in the Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 7 of Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans - 16.7 Mgy within 5.0Å range:
|
Reference:
Y.Fukuda,
K.M.Tse,
M.Suzuki,
K.Diederichs,
K.Hirata,
T.Nakane,
M.Sugahara,
E.Nango,
K.Tono,
Y.Joti,
T.Kameshima,
C.Song,
T.Hatsui,
M.Yabashi,
O.Nureki,
H.Matsumura,
T.Inoue,
S.Iwata,
E.Mizohata.
Redox-Coupled Structural Changes in Nitrite Reductase Revealed By Serial Femtosecond and Microfocus Crystallography J.Biochem. V. 159 527 2016.
ISSN: ISSN 0021-924X
PubMed: 26769972
DOI: 10.1093/JB/MVV133
Page generated: Mon Jul 14 04:13:46 2025
ISSN: ISSN 0021-924X
PubMed: 26769972
DOI: 10.1093/JB/MVV133
Last articles
I in 4B9HI in 4AS2
I in 4AS5
I in 4AX2
I in 4ARR
I in 4AQ3
I in 4ARK
I in 4AP2
I in 4AIO
I in 4ANB