Copper »
PDB 2xv2-2z7w »
2yet »
Copper in PDB 2yet: Thermoascus GH61 Isozyme A
Enzymatic activity of Thermoascus GH61 Isozyme A
All present enzymatic activity of Thermoascus GH61 Isozyme A:
3.2.1.4;
3.2.1.4;
Protein crystallography data
The structure of Thermoascus GH61 Isozyme A, PDB code: 2yet
was solved by
H.Otten,
R.J.Quinlan,
M.D.Sweeney,
J.-C.N.Poulsen,
K.S.Johansen,
K.B.R.M.Krogh,
C.I.Joergensen,
M.Tovborg,
A.Anthonsen,
T.Tryfona,
C.P.Walter,
P.Dupree,
F.Xu,
G.J.Davies,
P.H.Walton,
L.Lo Leggio,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 12.33 / 1.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 37.404, 88.467, 70.324, 90.00, 103.41, 90.00 |
| R / Rfree (%) | 14.4 / 18.5 |
Copper Binding Sites:
The binding sites of Copper atom in the Thermoascus GH61 Isozyme A
(pdb code 2yet). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Thermoascus GH61 Isozyme A, PDB code: 2yet:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Thermoascus GH61 Isozyme A, PDB code: 2yet:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 2yet
Go back to
Copper binding site 1 out
of 2 in the Thermoascus GH61 Isozyme A
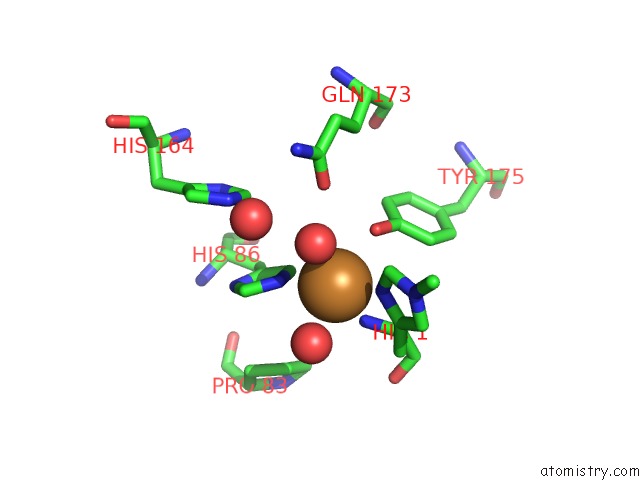
Mono view
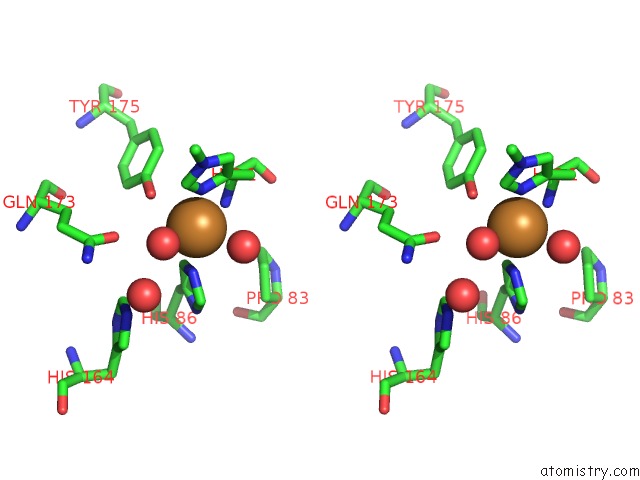
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Thermoascus GH61 Isozyme A within 5.0Å range:
|
Copper binding site 2 out of 2 in 2yet
Go back to
Copper binding site 2 out
of 2 in the Thermoascus GH61 Isozyme A
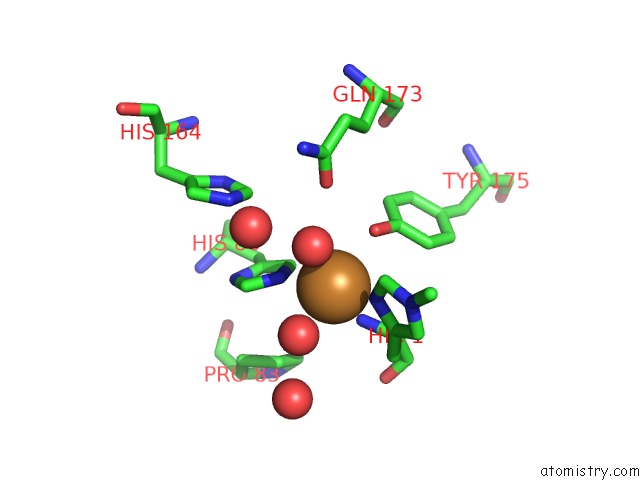
Mono view
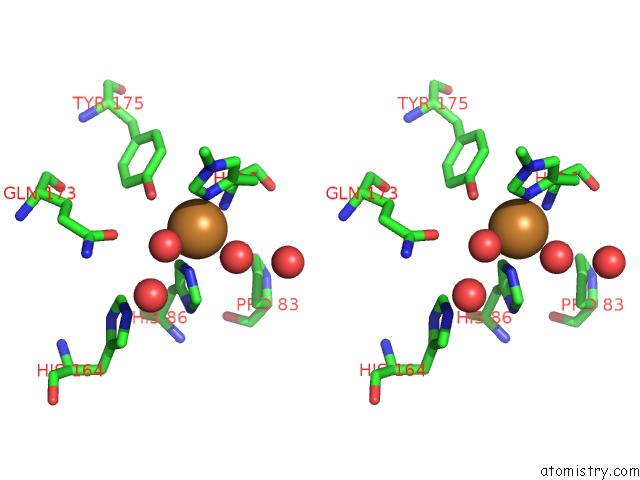
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Thermoascus GH61 Isozyme A within 5.0Å range:
|
Reference:
R.J.Quinlan,
M.D.Sweeney,
L.Lo Leggio,
H.Otten,
J.-C.N.Poulsen,
K.S.Johansen,
K.B.R.M.Krogh,
C.I.Jorgensen,
M.Tovborg,
A.Anthonsen,
T.Tryfona,
C.P.Walter,
P.Dupree,
F.Xu,
G.J.Davies,
P.H.Walton.
Insights Into the Oxidative Degradation of Cellulose By A Copper Metalloenzyme That Exploits Biomass Components. Proc.Natl.Acad.Sci.Usa V. 108 15079 2011.
ISSN: ISSN 0027-8424
PubMed: 21876164
DOI: 10.1073/PNAS.1105776108
Page generated: Mon Jul 14 01:42:31 2025
ISSN: ISSN 0027-8424
PubMed: 21876164
DOI: 10.1073/PNAS.1105776108
Last articles
K in 2VQWK in 2VQV
K in 2VQQ
K in 2VQO
K in 2VI5
K in 2VQM
K in 2VQJ
K in 2VPL
K in 2VLH
K in 2VLF