Copper »
PDB 1a2v-1baw »
1ba9 »
Copper in PDB 1ba9: The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures
Enzymatic activity of The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures
All present enzymatic activity of The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures:
1.15.1.1;
1.15.1.1;
Other elements in 1ba9:
The structure of The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures also contains other interesting chemical elements:
| Zinc | (Zn) | 36 atoms |
Copper Binding Sites:
The binding sites of Copper atom in the The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures
(pdb code 1ba9). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures, PDB code: 1ba9:
In total only one binding site of Copper was determined in the The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures, PDB code: 1ba9:
Copper binding site 1 out of 1 in 1ba9
Go back to
Copper binding site 1 out
of 1 in the The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures
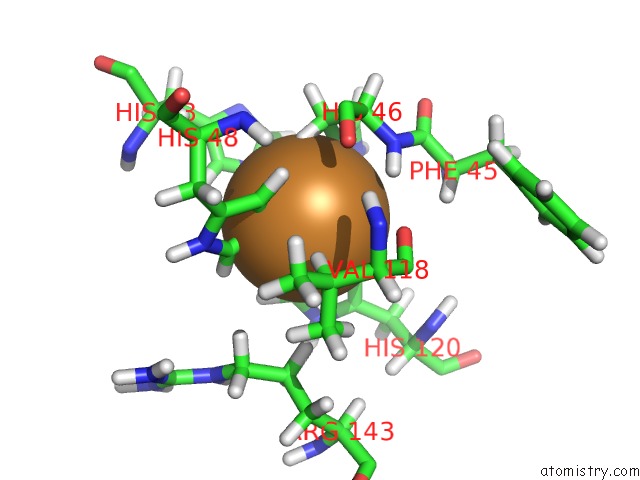
Mono view
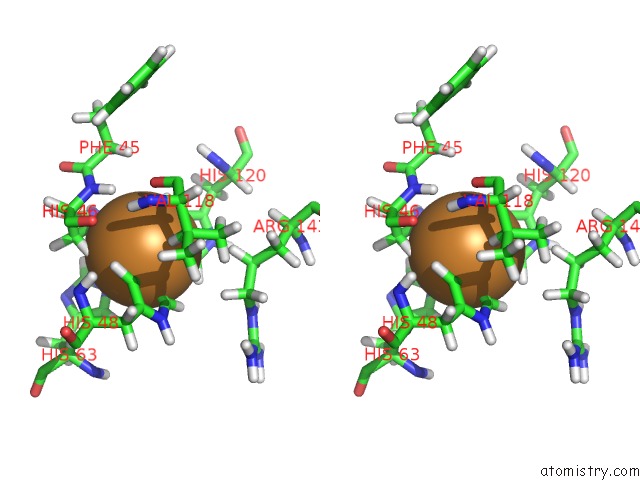
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of The Solution Structure of Reduced Monomeric Superoxide Dismutase, uc(Nmr), 36 Structures within 5.0Å range:
|
Reference:
L.Banci,
M.Benedetto,
I.Bertini,
R.Del Conte,
M.Piccioli,
M.S.Viezzoli.
Solution Structure of Reduced Monomeric Q133M2 Copper, Zinc Superoxide Dismutase (Sod). Why Is Sod A Dimeric Enzyme?. Biochemistry V. 37 11780 1998.
ISSN: ISSN 0006-2960
PubMed: 9718300
DOI: 10.1021/BI9803473
Page generated: Sun Jul 13 23:32:05 2025
ISSN: ISSN 0006-2960
PubMed: 9718300
DOI: 10.1021/BI9803473
Last articles
Hg in 3GFHHg in 3FW0
Hg in 3FN8
Hg in 3F8E
Hg in 3FFP
Hg in 3F4X
Hg in 3F2F
Hg in 3F2H
Hg in 3F0P
Hg in 3D8D