Copper »
PDB 8yk7-9fdl »
9fdl »
Copper in PDB 9fdl: Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F)
Protein crystallography data
The structure of Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F), PDB code: 9fdl
was solved by
C.Kosinas,
M.Dimarogona,
E.Topakas,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 131.54 / 2.33 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 55.891, 112.293, 262.381, 90, 90, 90 |
| R / Rfree (%) | 16.2 / 20.4 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F)
(pdb code 9fdl). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 3 binding sites of Copper where determined in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F), PDB code: 9fdl:
Jump to Copper binding site number: 1; 2; 3;
In total 3 binding sites of Copper where determined in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F), PDB code: 9fdl:
Jump to Copper binding site number: 1; 2; 3;
Copper binding site 1 out of 3 in 9fdl
Go back to
Copper binding site 1 out
of 3 in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F)
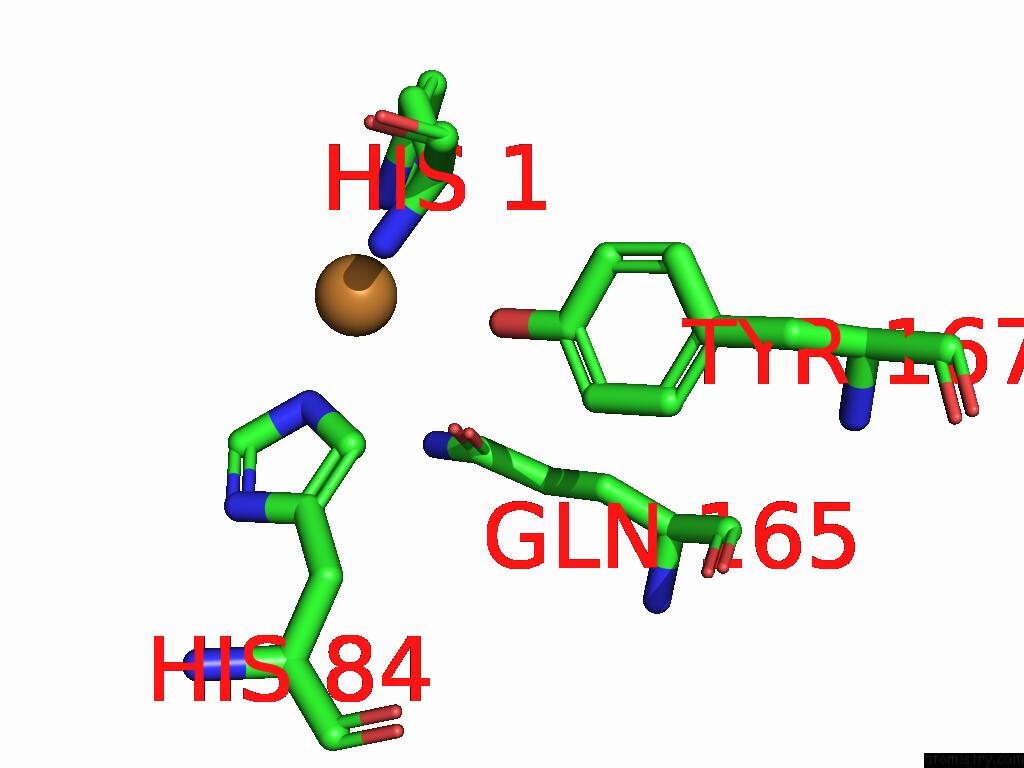
Mono view
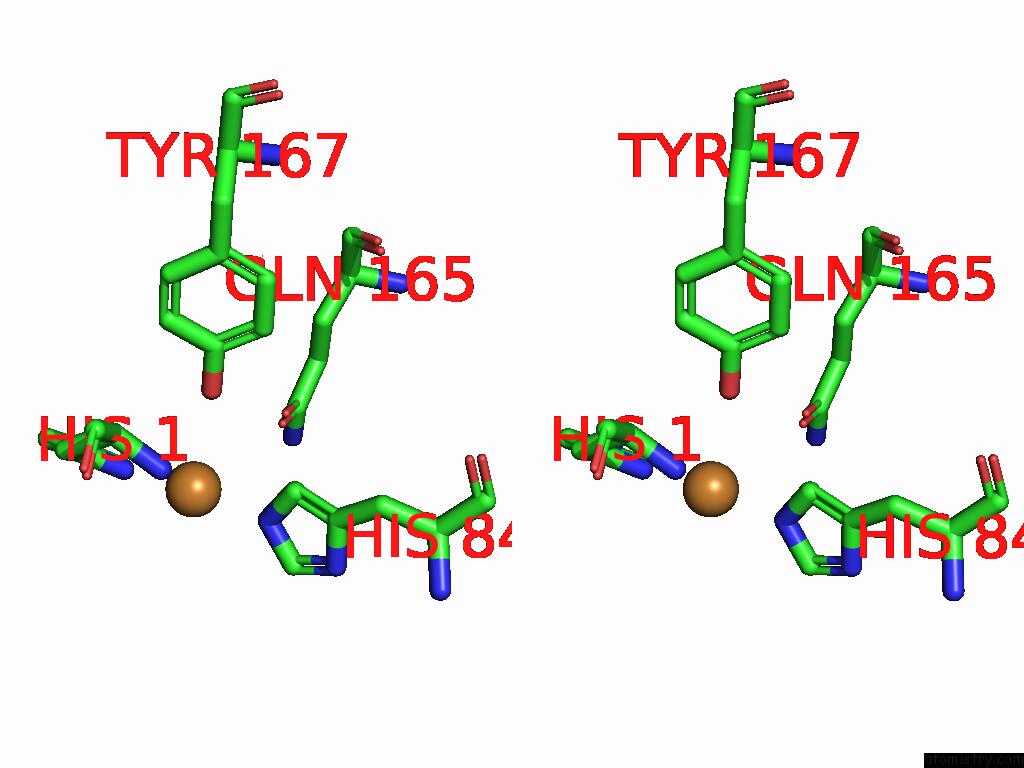
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F) within 5.0Å range:
|
Copper binding site 2 out of 3 in 9fdl
Go back to
Copper binding site 2 out
of 3 in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F)
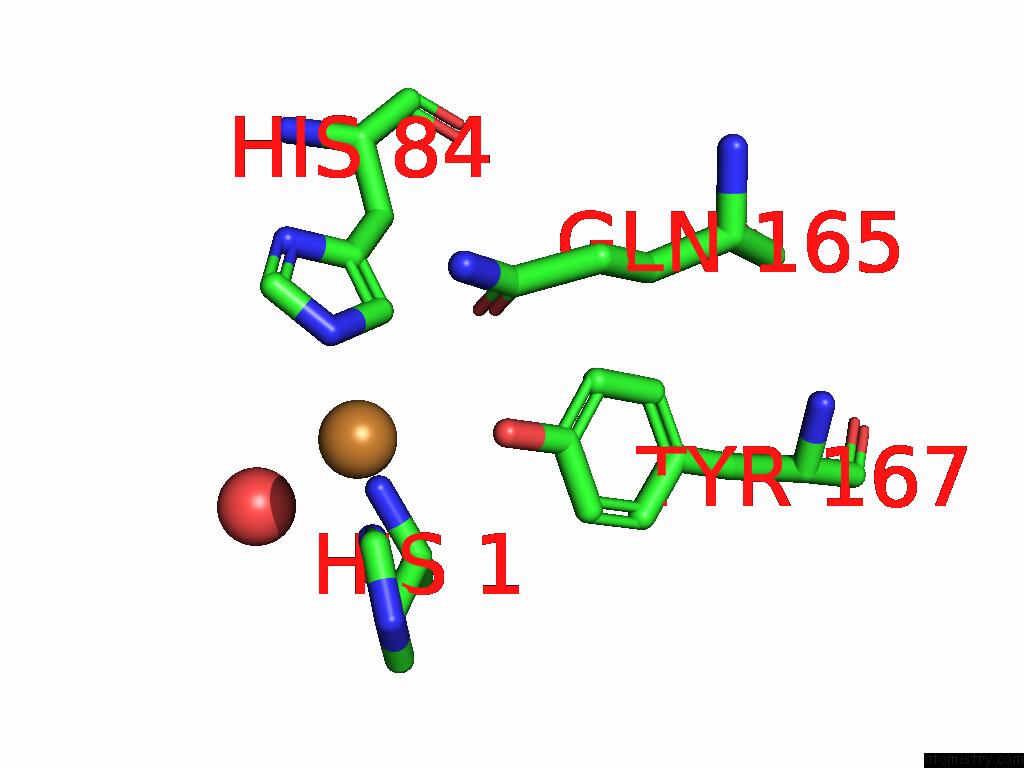
Mono view
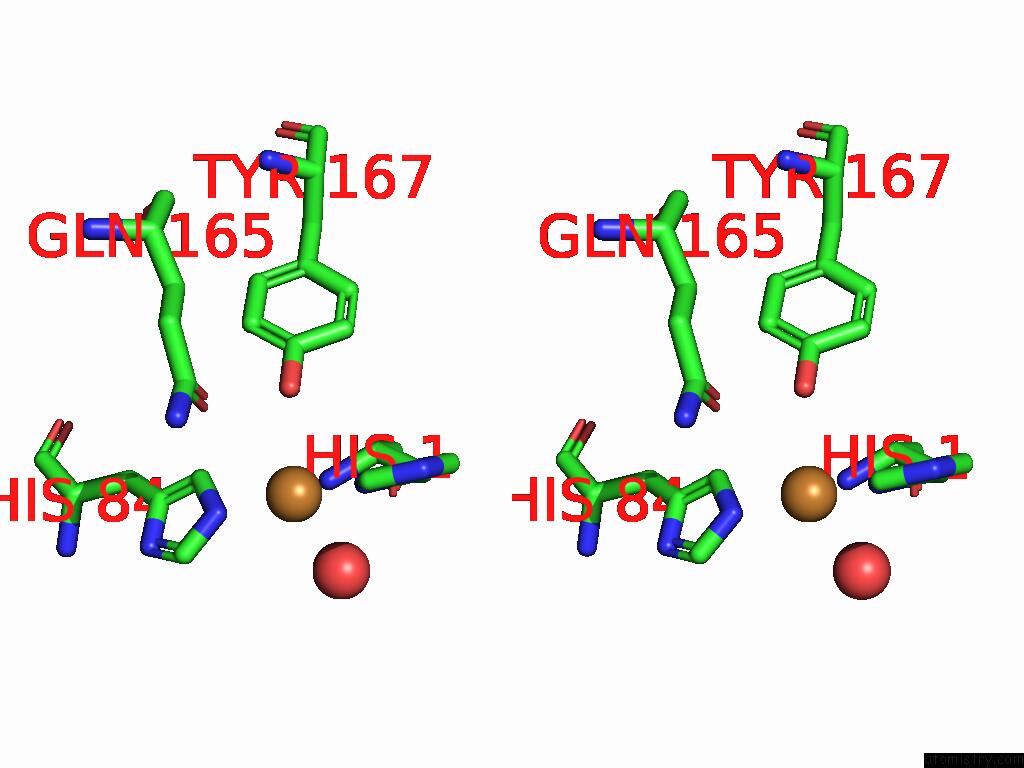
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F) within 5.0Å range:
|
Copper binding site 3 out of 3 in 9fdl
Go back to
Copper binding site 3 out
of 3 in the Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F)
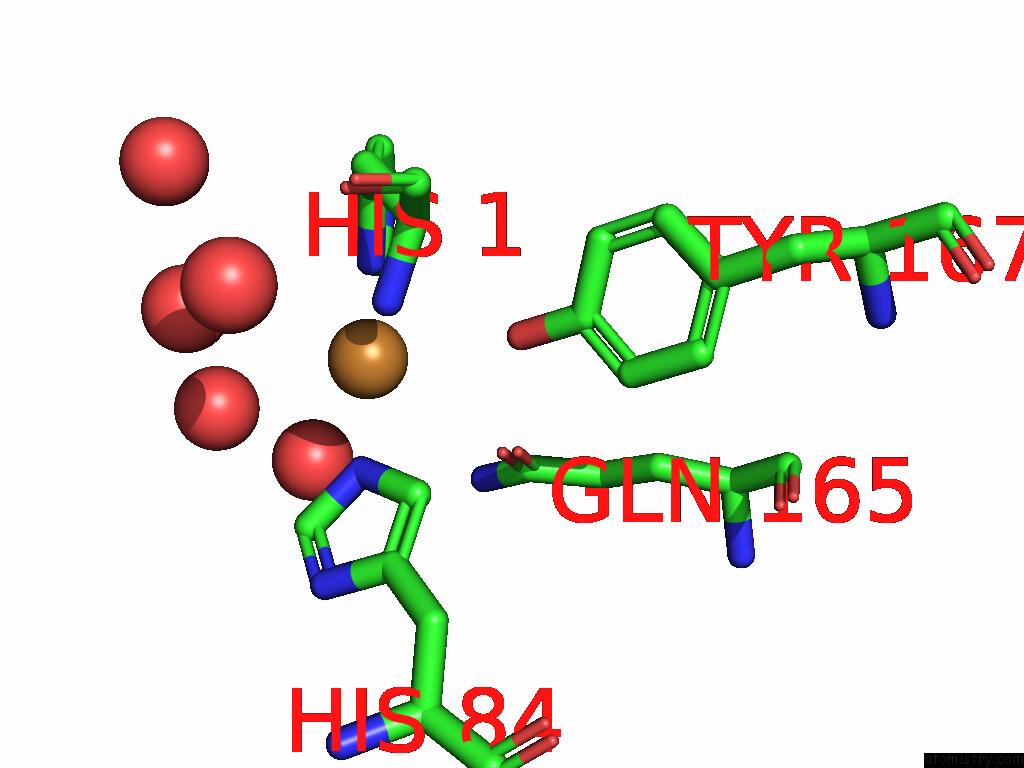
Mono view
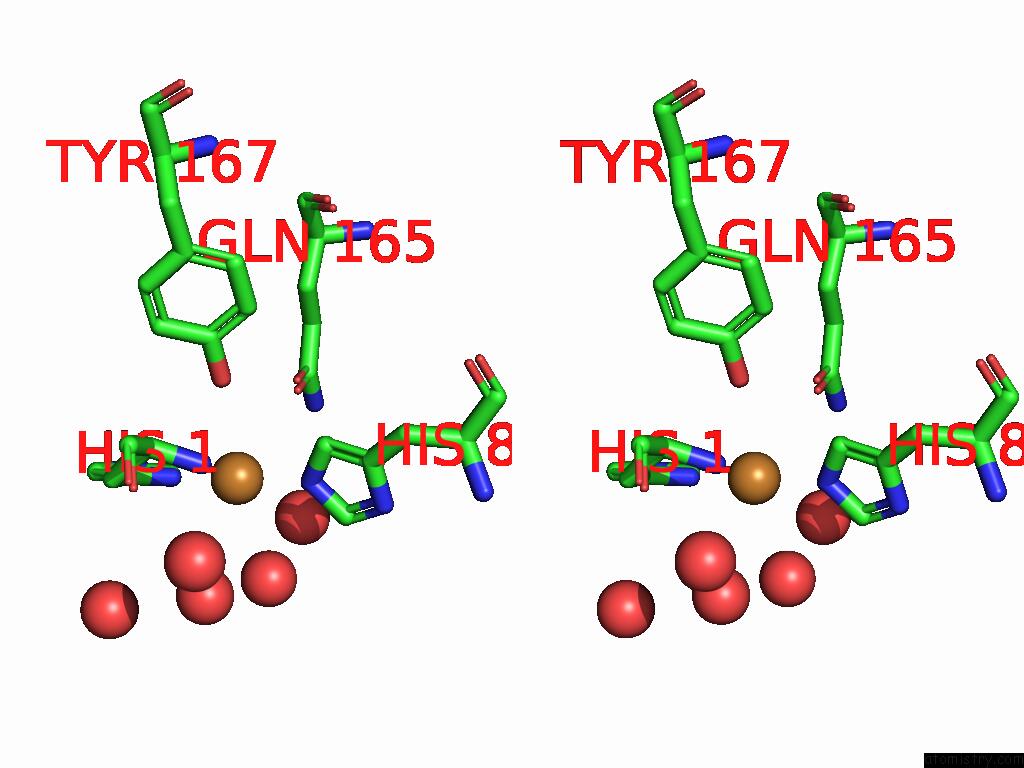
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 3 of Crystal Structure of the Catalytic Domain of An AA9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (TTLPMO9F) within 5.0Å range:
|
Reference:
C.Kosinas,
K.Chorozian,
M.Sandgren,
E.Topakas,
M.Dimarogona.
Mutational Study of A Lytic Polysaccharide Monooxygenase From Myceliophthora Thermophila (MTLPMO9F): Structural Insights Into Substrate Specificity and Regioselectivity. Int.J.Biol.Macromol. V. 288 38574 2024.
ISSN: ISSN 0141-8130
PubMed: 39662565
DOI: 10.1016/J.IJBIOMAC.2024.138574
Page generated: Sat Feb 8 17:42:00 2025
ISSN: ISSN 0141-8130
PubMed: 39662565
DOI: 10.1016/J.IJBIOMAC.2024.138574
Last articles
Cl in 3DCXCl in 3DCU
Cl in 3D53
Cl in 3DCT
Cl in 3DCJ
Cl in 3DBF
Cl in 3DCC
Cl in 3DBE
Cl in 3DAQ
Cl in 3DBD