Copper »
PDB 8prq-8ryv »
8qya »
Copper in PDB 8qya: J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation
Protein crystallography data
The structure of J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation, PDB code: 8qya
was solved by
S.F.Marino,
O.Daumke,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 21.76 / 2.72 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 137.266, 55.371, 69.486, 90, 108.01, 90 |
| R / Rfree (%) | 24.4 / 29.7 |
Copper Binding Sites:
The binding sites of Copper atom in the J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation
(pdb code 8qya). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation, PDB code: 8qya:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation, PDB code: 8qya:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 8qya
Go back to
Copper binding site 1 out
of 2 in the J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation
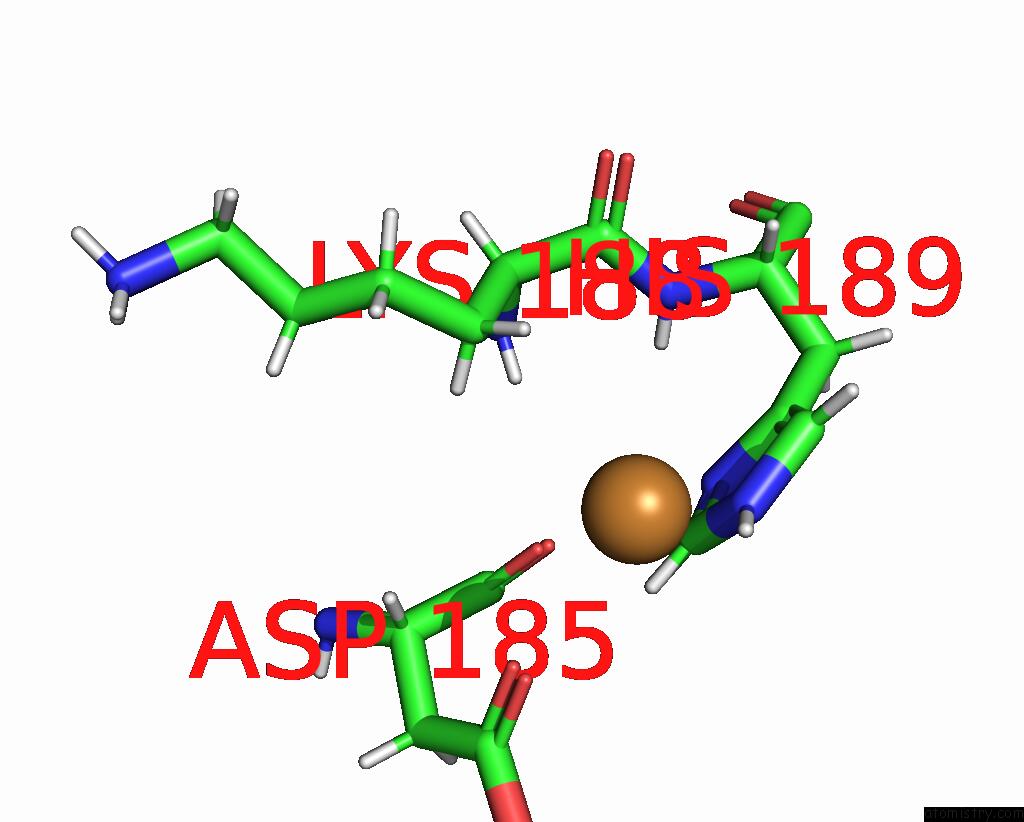
Mono view
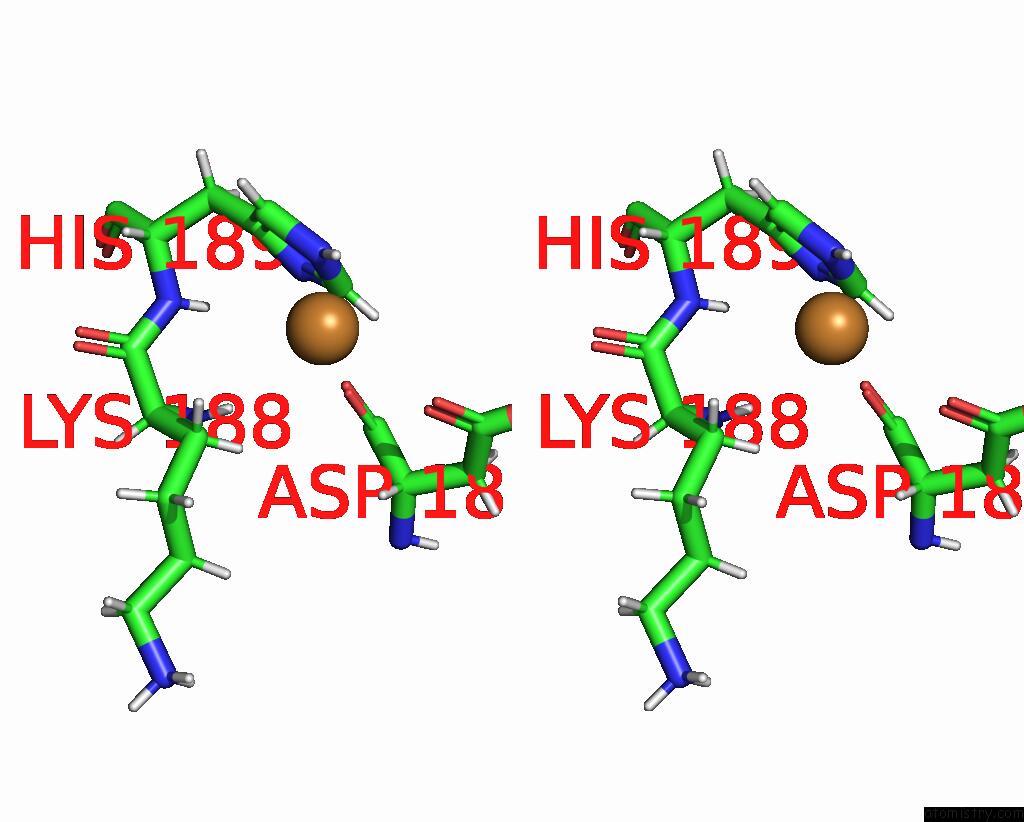
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation within 5.0Å range:
|
Copper binding site 2 out of 2 in 8qya
Go back to
Copper binding site 2 out
of 2 in the J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation
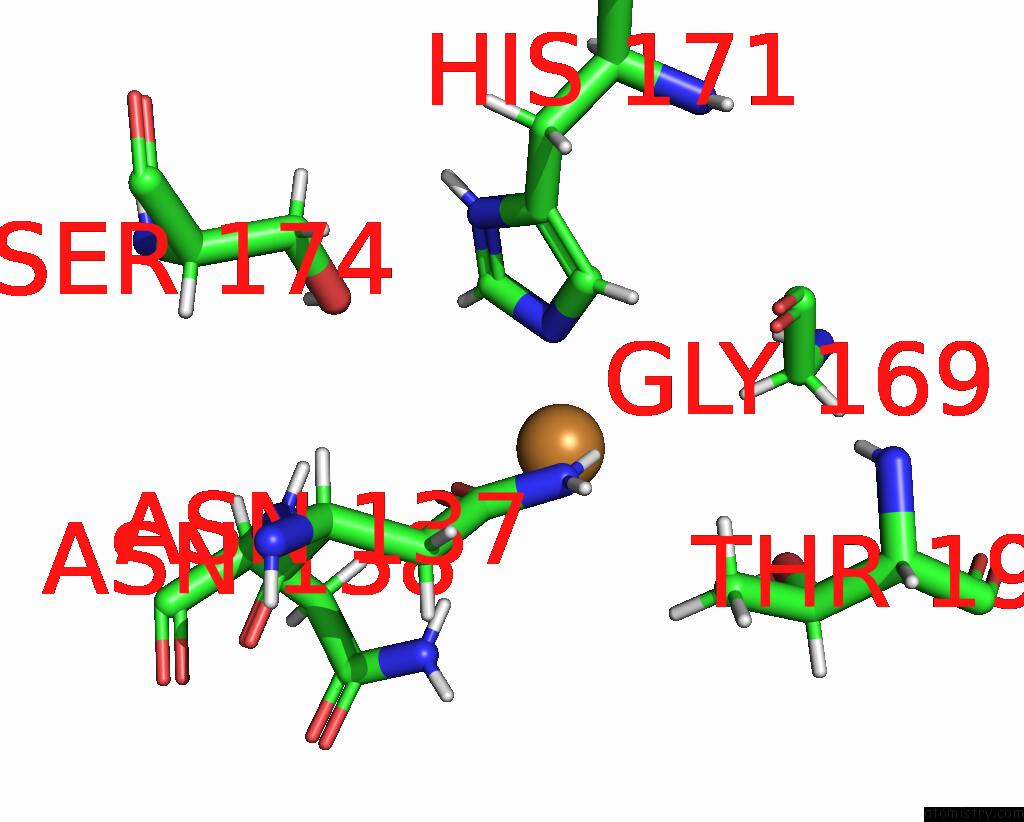
Mono view
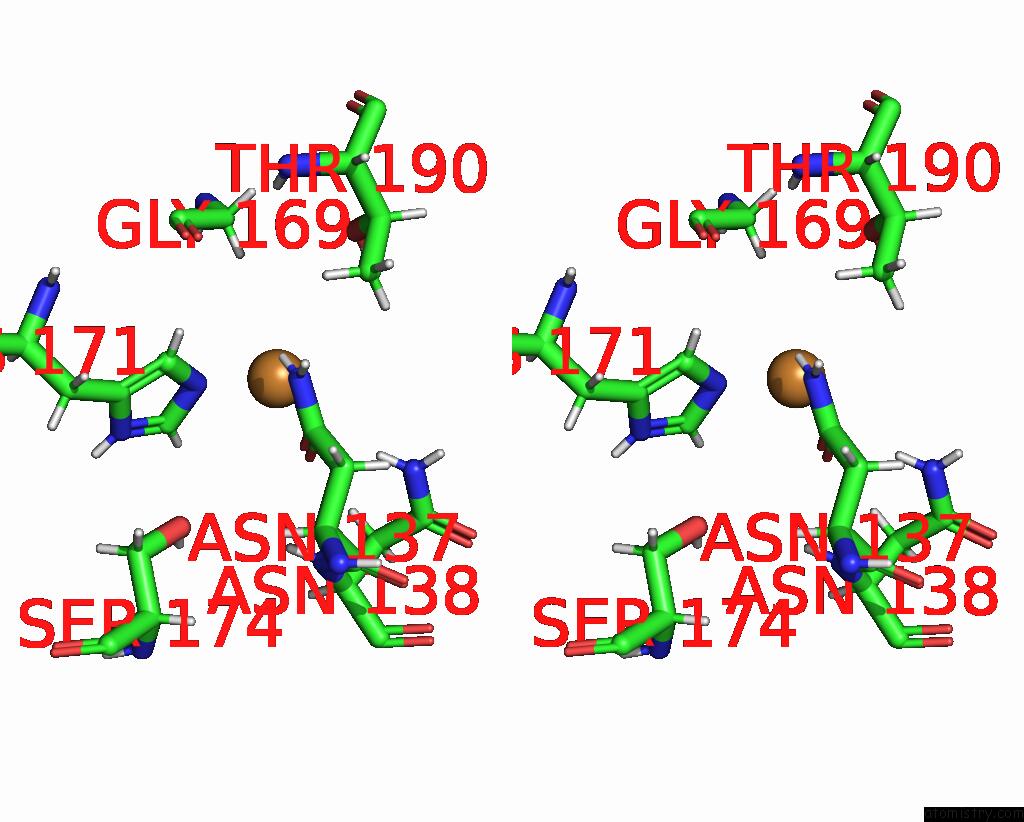
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of J22.9-Fny, Fully Humanized, Cdr Optimized Fab Fragment Based on Chimeric J22.9-XI Igg Against Bcma; with Vh CDR2 Glycosylation within 5.0Å range:
|
Reference:
S.F.Marino,
O.Daumke.
Structure-Based Humanization of A Therapeutic Antibody For Multiple Myeloma To Be Published.
Page generated: Mon Jul 14 09:16:54 2025
Last articles
F in 7LQUF in 7LPF
F in 7LOE
F in 7LOM
F in 7LOD
F in 7LOC
F in 7LO6
F in 7LOA
F in 7LOB
F in 7LK1