Copper »
PDB 8prq-8s2q »
8qgf »
Copper in PDB 8qgf: Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
Protein crystallography data
The structure of Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii, PDB code: 8qgf
was solved by
N.Petchyam,
S.Antonyuk,
S.S.Hasnain,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.66 / 1.32 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 127.952, 127.952, 86.249, 90, 90, 120 |
| R / Rfree (%) | 13.4 / 16.2 |
Other elements in 8qgf:
The structure of Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii also contains other interesting chemical elements:
| Iron | (Fe) | 1 atom |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
(pdb code 8qgf). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii, PDB code: 8qgf:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii, PDB code: 8qgf:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 8qgf
Go back to
Copper binding site 1 out
of 2 in the Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
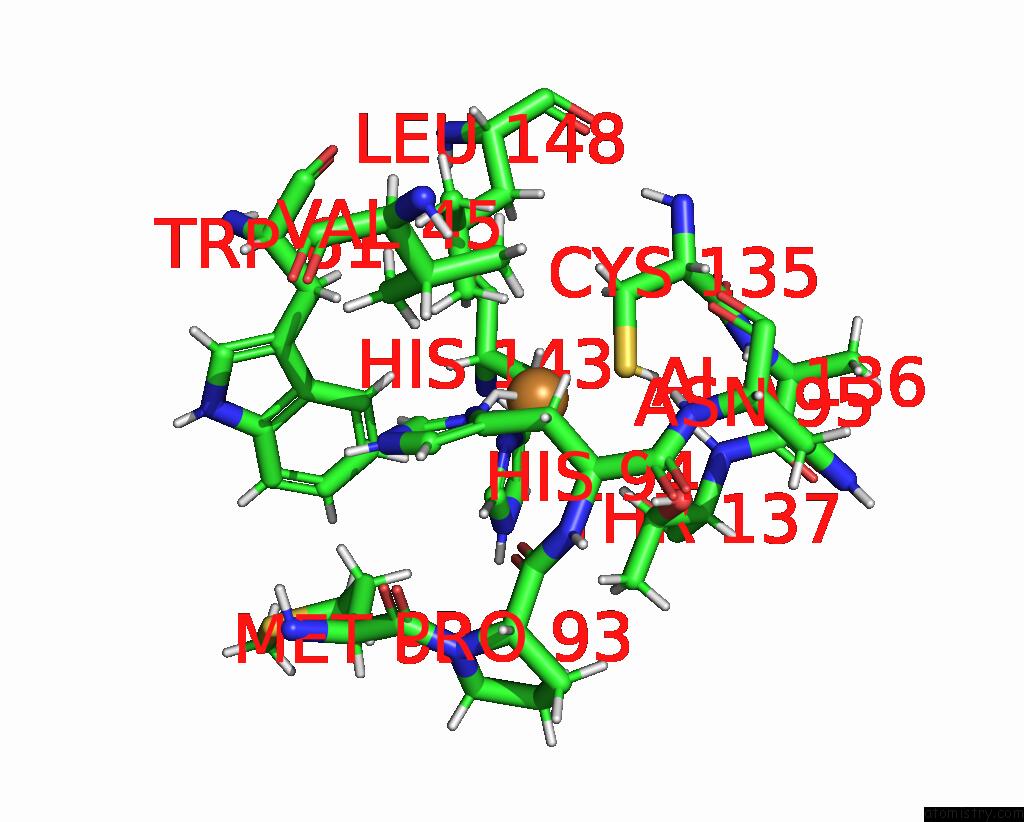
Mono view
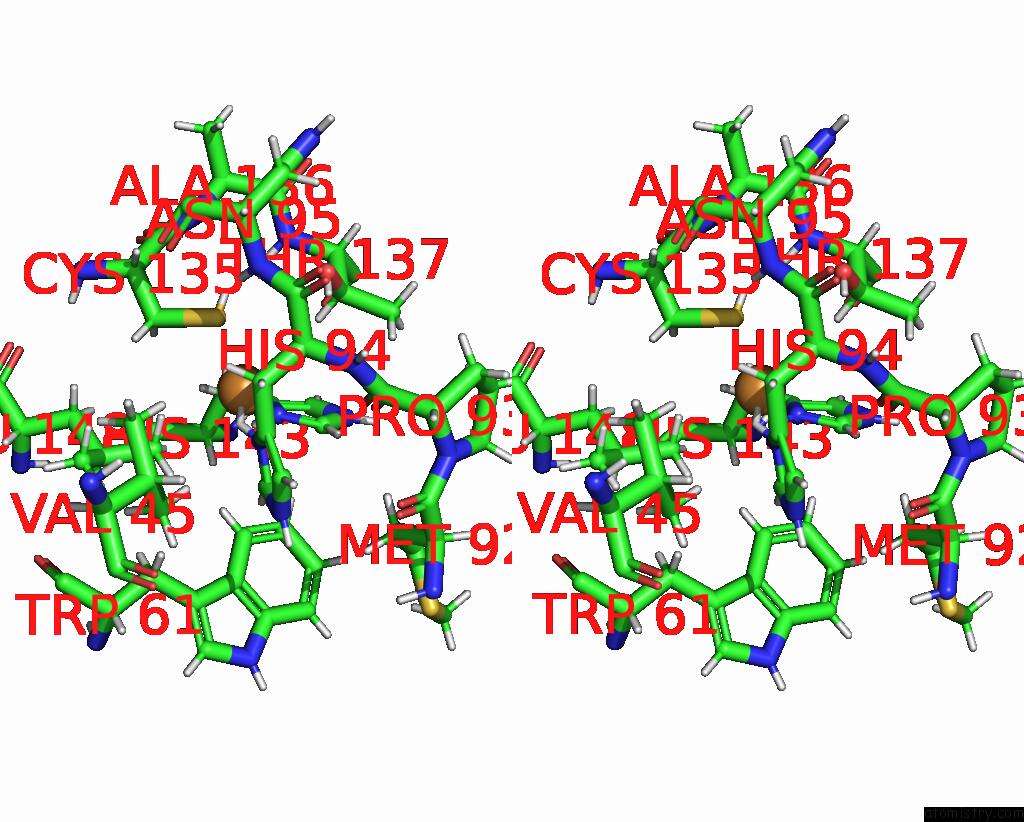
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii within 5.0Å range:
|
Copper binding site 2 out of 2 in 8qgf
Go back to
Copper binding site 2 out
of 2 in the Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii
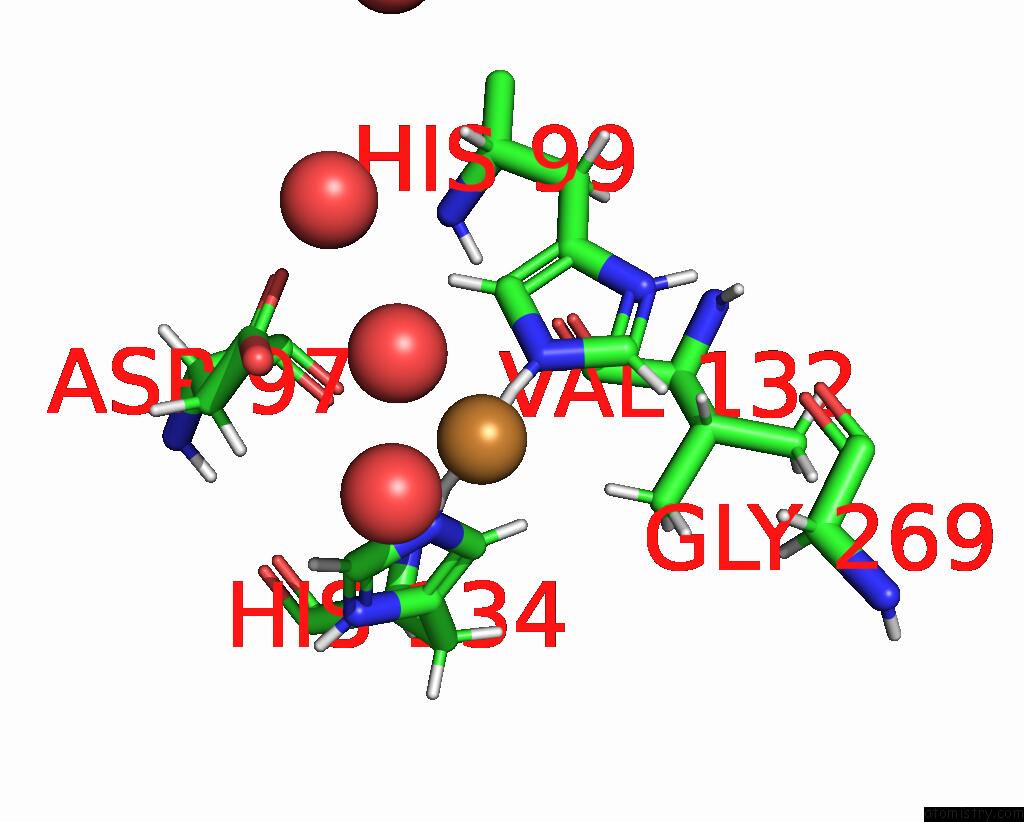
Mono view
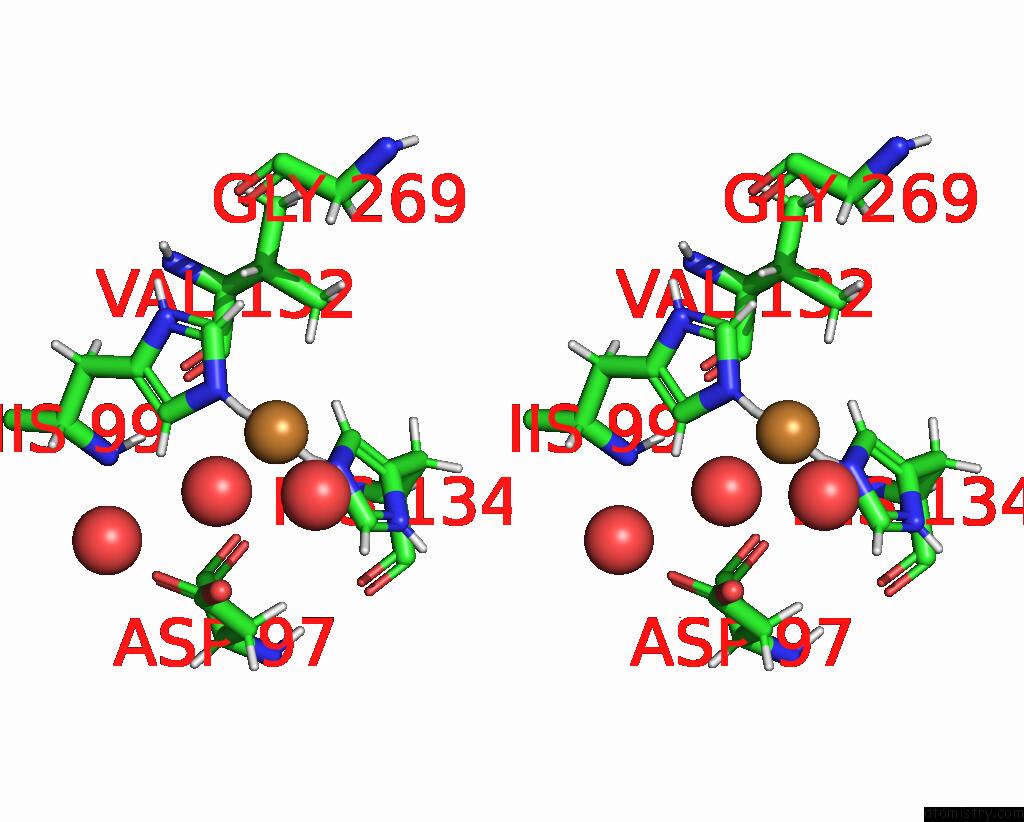
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of As-Isolated M148L Mutant of Three-Domain Heme-Cu Nitrite Reductase From Ralstonia Pickettii within 5.0Å range:
|
Reference:
N.Petchyam,
S.Antonyuk,
S.S.Hasnain.
Structural Studies of Haem Three-Domain Copper Nitrite Reductase Mutants From Ralstonia Pickettii To Be Published.
Page generated: Sat Sep 28 19:55:46 2024
Last articles
Cl in 5SDKCl in 5SDI
Cl in 5SDJ
Cl in 5SDH
Cl in 5SDD
Cl in 5SDE
Cl in 5SDF
Cl in 5SDG
Cl in 5SDC
Cl in 5SD6