Copper »
PDB 7s1f-7xmb »
7wno »
Copper in PDB 7wno: Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data.
Enzymatic activity of Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data.
All present enzymatic activity of Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data.:
1.4.3.21;
1.4.3.21;
Copper Binding Sites:
The binding sites of Copper atom in the Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data.
(pdb code 7wno). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data., PDB code: 7wno:
In total only one binding site of Copper was determined in the Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data., PDB code: 7wno:
Copper binding site 1 out of 1 in 7wno
Go back to
Copper binding site 1 out
of 1 in the Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data.

Mono view
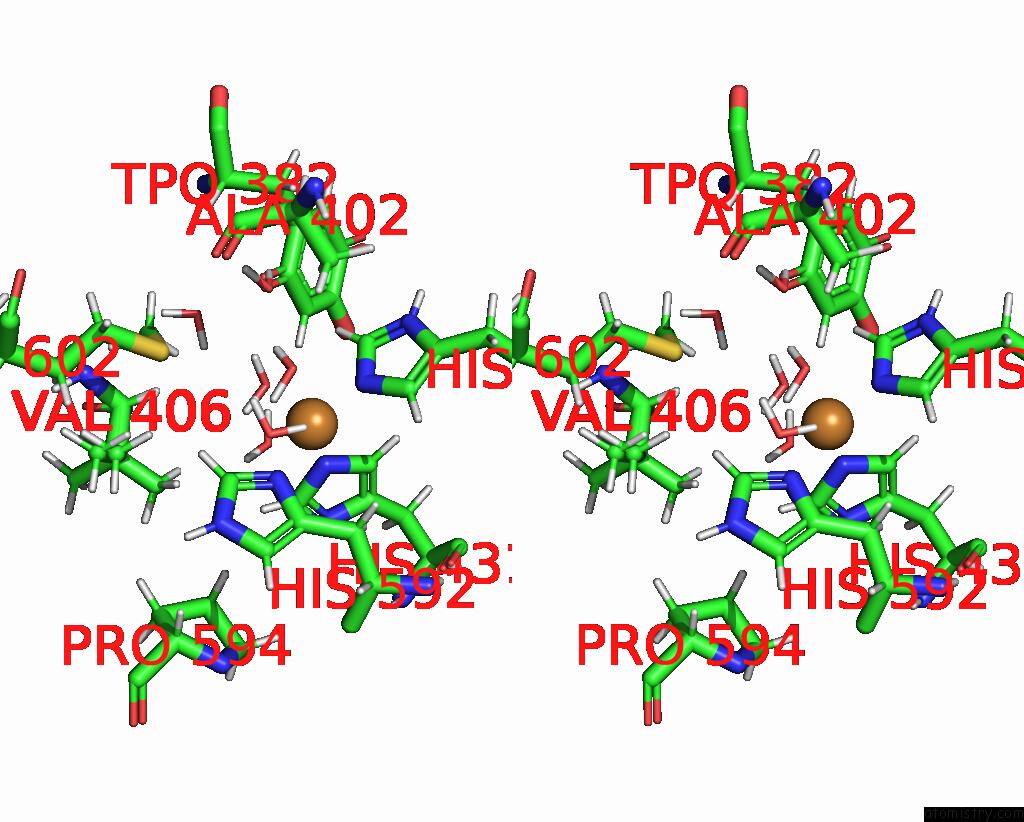
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystallographic Structure of Copper Amine Oxidase From Arthrobacter Glibiformis at Pd 7.4 Determined By Only Neutron Diffraction Data. within 5.0Å range:
|
Reference:
T.Murakawa,
K.Kurihara,
M.Adachi,
K.Kusaka,
K.Tanizawa,
T.Okajima.
Re-Evaluation of Protein Neutron Crystallography with and Without X-Ray/Neutron Joint Refinement. Iucrj V. 9 342 2022.
ISSN: ESSN 2052-2525
PubMed: 35546796
DOI: 10.1107/S2052252522003657
Page generated: Wed Jul 31 09:21:11 2024
ISSN: ESSN 2052-2525
PubMed: 35546796
DOI: 10.1107/S2052252522003657
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW