Copper »
PDB 7ev9-7o3c »
7n9z »
Copper in PDB 7n9z: E. Coli Cytochrome BO3 in Msp Nanodisc
Other elements in 7n9z:
The structure of E. Coli Cytochrome BO3 in Msp Nanodisc also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
| Iron | (Fe) | 2 atoms |
Copper Binding Sites:
The binding sites of Copper atom in the E. Coli Cytochrome BO3 in Msp Nanodisc
(pdb code 7n9z). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the E. Coli Cytochrome BO3 in Msp Nanodisc, PDB code: 7n9z:
In total only one binding site of Copper was determined in the E. Coli Cytochrome BO3 in Msp Nanodisc, PDB code: 7n9z:
Copper binding site 1 out of 1 in 7n9z
Go back to
Copper binding site 1 out
of 1 in the E. Coli Cytochrome BO3 in Msp Nanodisc
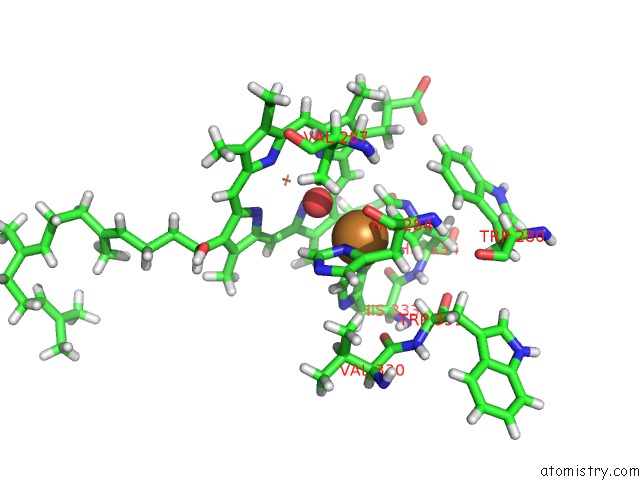
Mono view
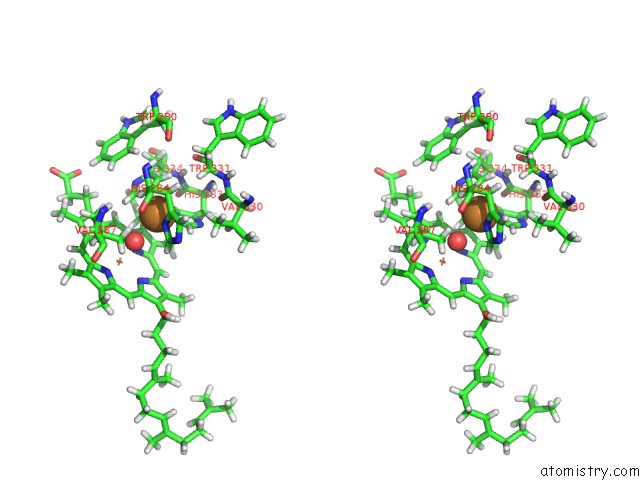
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of E. Coli Cytochrome BO3 in Msp Nanodisc within 5.0Å range:
|
Reference:
J.Li,
L.Han,
F.Vallese,
Z.Ding,
S.K.Choi,
S.Hong,
Y.Luo,
B.Liu,
C.K.Chan,
E.Tajkhorshid,
J.Zhu,
O.Clarke,
K.Zhang,
R.Gennis.
Cryo-Em Structures of Escherichia Coli Cytochrome Bo 3 Reveal Bound Phospholipids and Ubiquinone-8 in A Dynamic Substrate Binding Site. Proc.Natl.Acad.Sci.Usa V. 118 2021.
ISSN: ESSN 1091-6490
PubMed: 34417297
DOI: 10.1073/PNAS.2106750118
Page generated: Mon Jul 14 08:07:47 2025
ISSN: ESSN 1091-6490
PubMed: 34417297
DOI: 10.1073/PNAS.2106750118
Last articles
F in 7LGKF in 7LG8
F in 7LD3
F in 7LCR
F in 7LCM
F in 7LCO
F in 7LCK
F in 7LCJ
F in 7LCI
F in 7L9Y