Copper »
PDB 5zpp-6fja »
6br4 »
Copper in PDB 6br4: Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine
Protein crystallography data
The structure of Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine, PDB code: 6br4
was solved by
B.Heras,
M.Totsika,
J.J.Paxman,
G.Wang,
M.J.Scanlon,
J.L.Martin,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 52.84 / 1.99 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.224, 63.937, 74.510, 90.00, 126.15, 90.00 |
| R / Rfree (%) | 16.8 / 21.5 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine
(pdb code 6br4). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine, PDB code: 6br4:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine, PDB code: 6br4:
Jump to Copper binding site number: 1; 2;
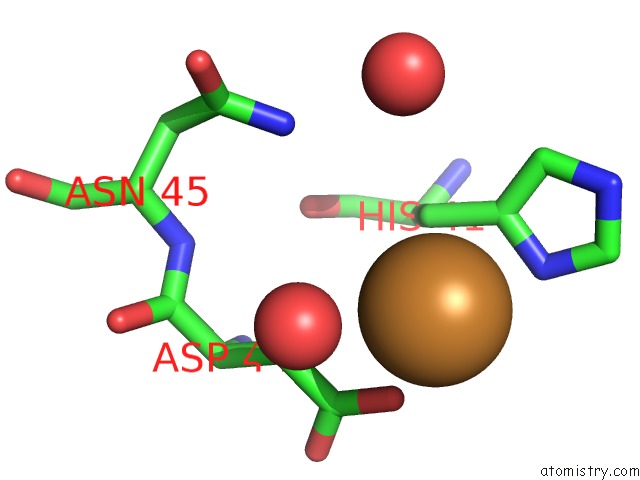
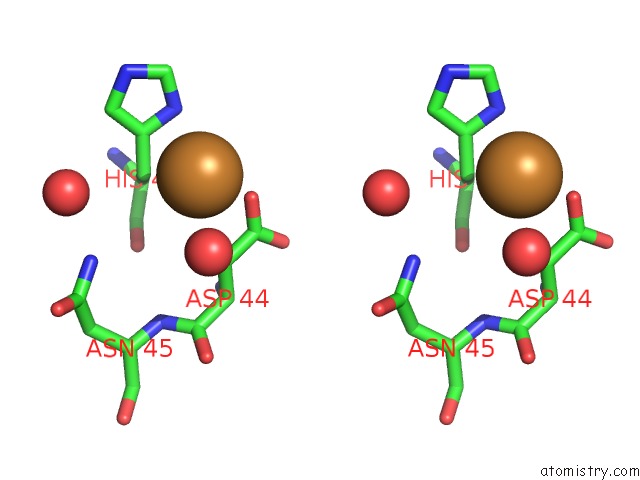
Copper binding site 1 out of 2 in 6br4
Go back to
Copper binding site 1 out
of 2 in the Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine within 5.0Å range:
|
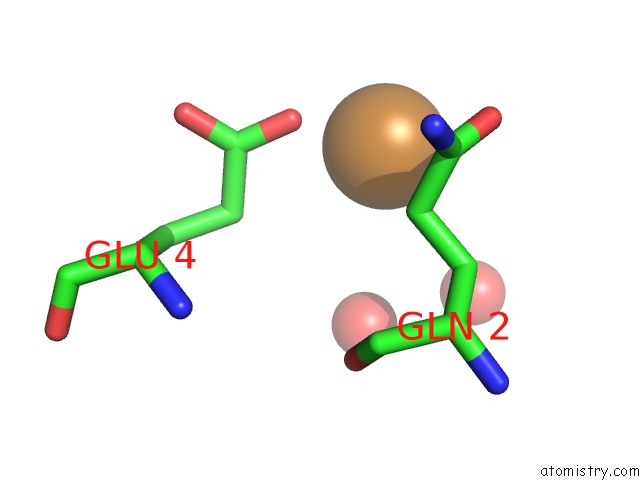
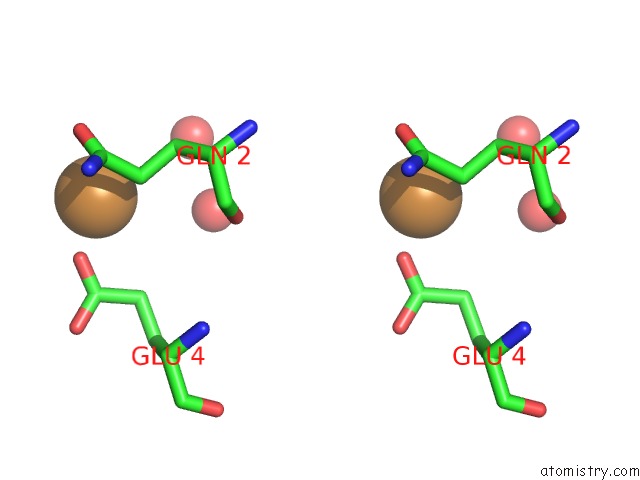
Copper binding site 2 out of 2 in 6br4
Go back to
Copper binding site 2 out
of 2 in the Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of Escherichia Coli Dsba in Complex with {N}-Methyl- 1-(3-Thiophen-2-Ylphenyl)Methanamine within 5.0Å range:
|
Reference:
M.Totsika,
D.Vagenas,
J.J.Paxman,
G.Wang,
R.Dhouib,
P.Sharma,
J.L.Martin,
M.J.Scanlon,
B.Heras.
Inhibition of Diverse Dsba Enzymes in Multi-Dsba Encoding Pathogens. Antioxid. Redox Signal. V. 29 653 2018.
ISSN: ESSN 1557-7716
PubMed: 29237285
DOI: 10.1089/ARS.2017.7104
Page generated: Mon Jul 14 05:54:45 2025
ISSN: ESSN 1557-7716
PubMed: 29237285
DOI: 10.1089/ARS.2017.7104
Last articles
Fe in 1V0HFe in 1V07
Fe in 1UWV
Fe in 1UX9
Fe in 1UZW
Fe in 1UYU
Fe in 1UWM
Fe in 1UX8
Fe in 1UPX
Fe in 1UVY