Copper »
PDB 5i26-5luf »
5i6p »
Copper in PDB 5i6p: Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy
Enzymatic activity of Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy
All present enzymatic activity of Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy:
1.7.2.1;
1.7.2.1;
Protein crystallography data
The structure of Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy, PDB code: 5i6p
was solved by
S.Horrell,
M.A.Hough,
R.W.Strange,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 42.60 / 1.56 |
| Space group | P 21 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 95.303, 95.303, 95.303, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.9 / 19.5 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy
(pdb code 5i6p). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy, PDB code: 5i6p:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy, PDB code: 5i6p:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 5i6p
Go back to
Copper binding site 1 out
of 2 in the Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy
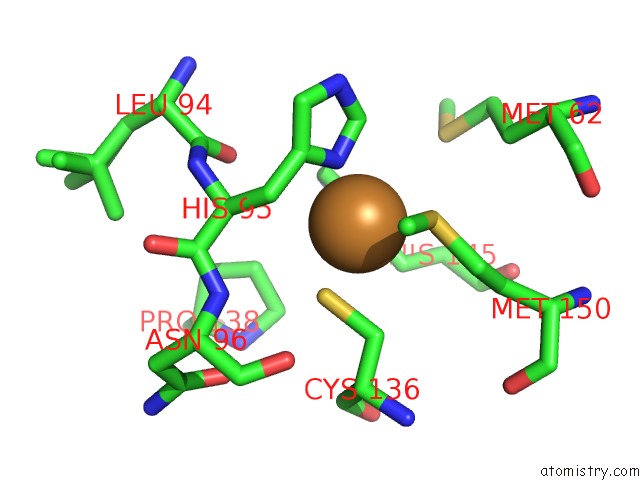
Mono view
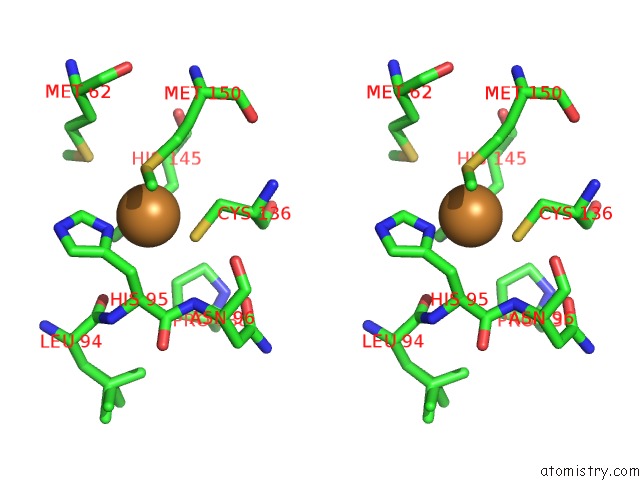
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy within 5.0Å range:
|
Copper binding site 2 out of 2 in 5i6p
Go back to
Copper binding site 2 out
of 2 in the Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy
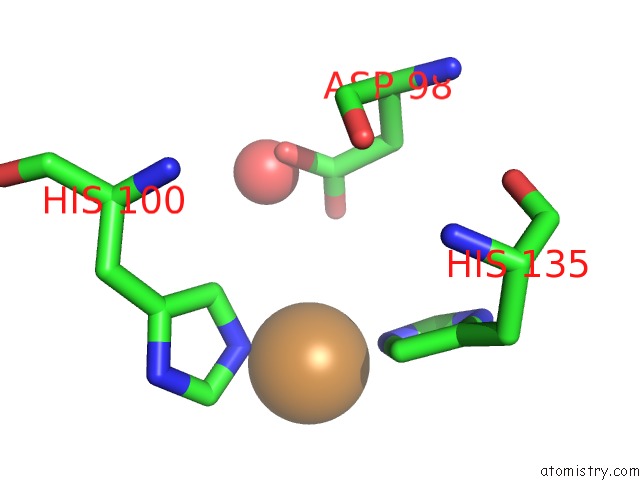
Mono view
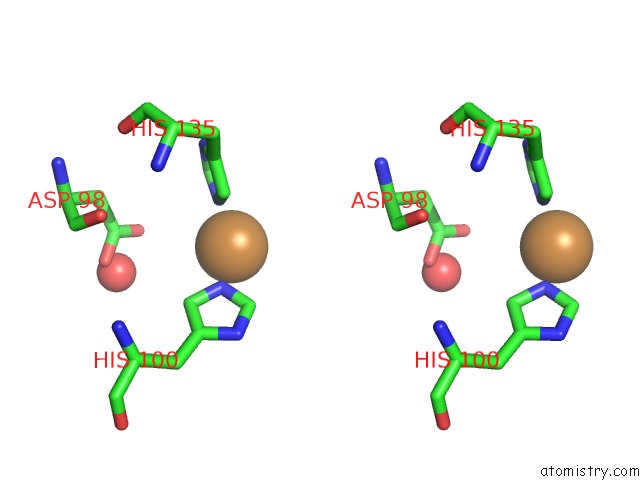
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of Copper Nitrite Reductase at 100K After 27.60 Mgy within 5.0Å range:
|
Reference:
S.Horrell,
S.V.Antonyuk,
R.R.Eady,
S.S.Hasnain,
M.A.Hough,
R.W.Strange.
Serial Crystallography Captures Enzyme Catalysis in Copper Nitrite Reductase at Atomic Resolution From One Crystal. Iucrj V. 3 271 2016.
ISSN: ESSN 2052-2525
PubMed: 27437114
DOI: 10.1107/S205225251600823X
Page generated: Mon Jul 14 04:43:00 2025
ISSN: ESSN 2052-2525
PubMed: 27437114
DOI: 10.1107/S205225251600823X
Last articles
Fe in 1PO5Fe in 1PMB
Fe in 1PL3
Fe in 1PM1
Fe in 1PJ1
Fe in 1PKF
Fe in 1PJ0
Fe in 1PIZ
Fe in 1PIY
Fe in 1PIJ