Copper »
PDB 4ysu-5c92 »
4z0z »
Copper in PDB 4z0z: Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
Protein crystallography data
The structure of Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %, PDB code: 4z0z
was solved by
C.Molitor,
S.G.Mauracher,
A.Rompel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.10 / 1.60 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.530, 183.520, 78.090, 90.00, 94.50, 90.00 |
| R / Rfree (%) | 16.5 / 20 |
Copper Binding Sites:
The binding sites of Copper atom in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
(pdb code 4z0z). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 4 binding sites of Copper where determined in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %, PDB code: 4z0z:
Jump to Copper binding site number: 1; 2; 3; 4;
In total 4 binding sites of Copper where determined in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %, PDB code: 4z0z:
Jump to Copper binding site number: 1; 2; 3; 4;
Copper binding site 1 out of 4 in 4z0z
Go back to
Copper binding site 1 out
of 4 in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
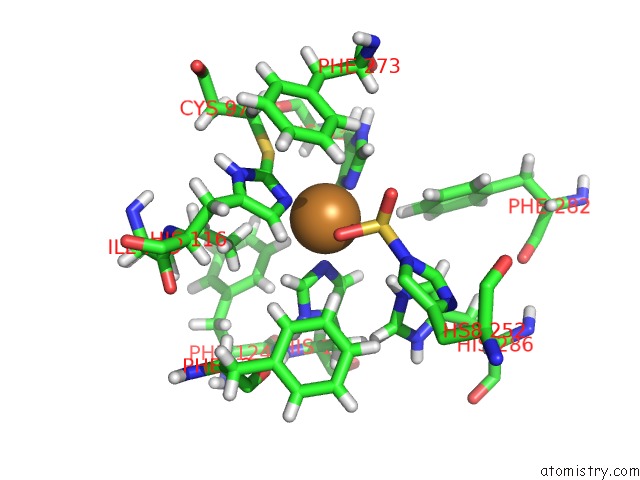
Mono view
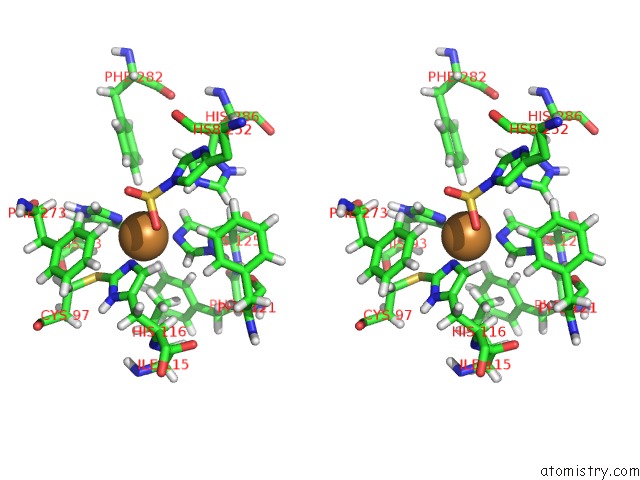
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 % within 5.0Å range:
|
Copper binding site 2 out of 4 in 4z0z
Go back to
Copper binding site 2 out
of 4 in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
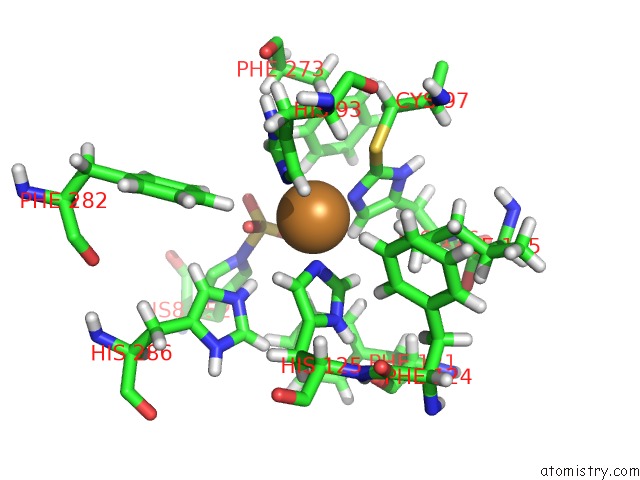
Mono view
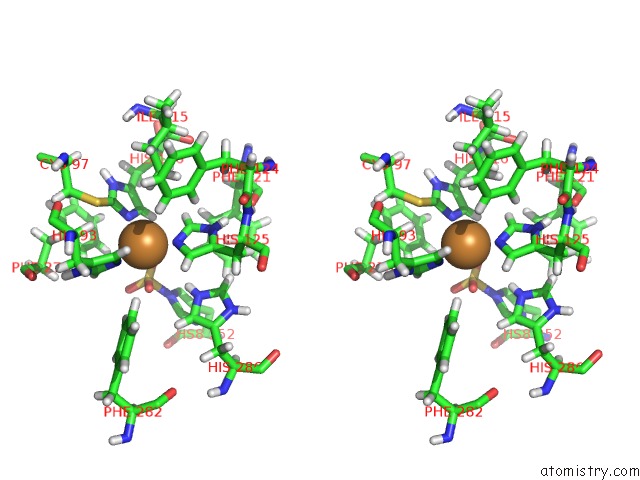
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 % within 5.0Å range:
|
Copper binding site 3 out of 4 in 4z0z
Go back to
Copper binding site 3 out
of 4 in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
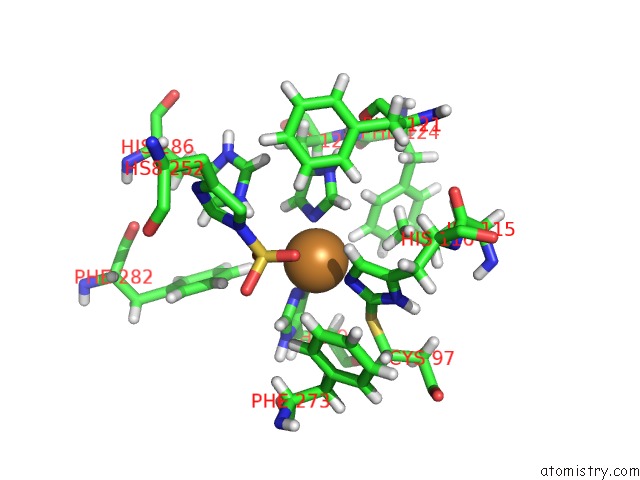
Mono view
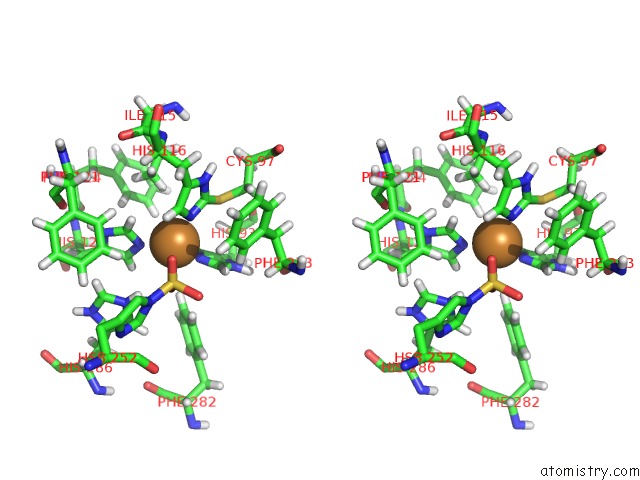
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 3 of Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 % within 5.0Å range:
|
Copper binding site 4 out of 4 in 4z0z
Go back to
Copper binding site 4 out
of 4 in the Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 %
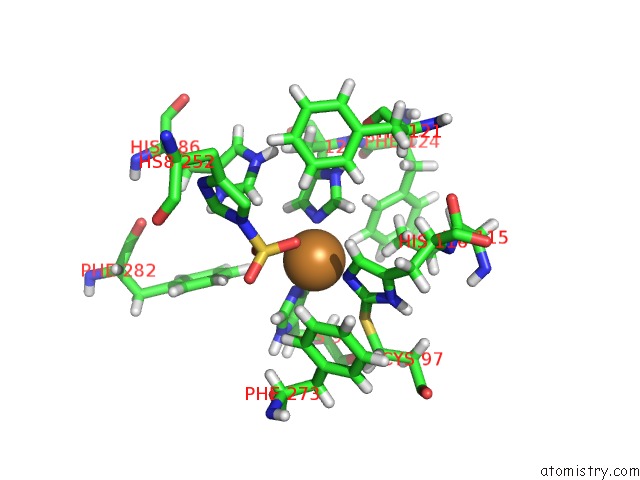
Mono view
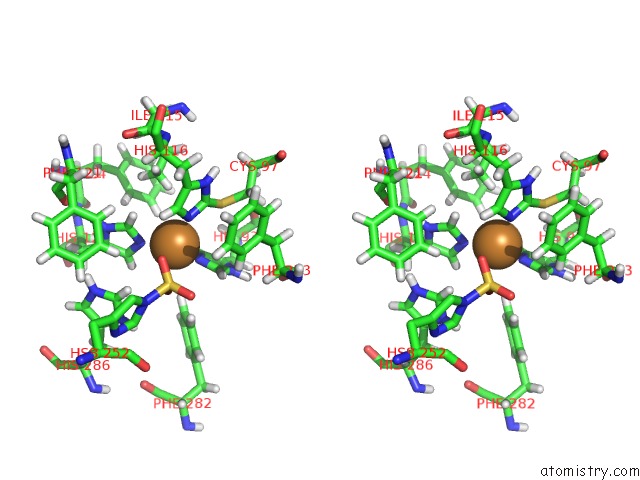
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 4 of Inactive Aurone Synthase (Polyphenol Oxidase) From Natural Source, Sulfohistidine ~ 90 % within 5.0Å range:
|
Reference:
C.Molitor,
S.G.Mauracher,
A.Rompel.
Aurone Synthase Is A Catechol Oxidase with Hydroxylase Activity and Provides Insights Into the Mechanism of Plant Polyphenol Oxidases. Proc.Natl.Acad.Sci.Usa V. 113 E1806 2016.
ISSN: ESSN 1091-6490
PubMed: 26976571
DOI: 10.1073/PNAS.1523575113
Page generated: Wed Jul 31 03:42:43 2024
ISSN: ESSN 1091-6490
PubMed: 26976571
DOI: 10.1073/PNAS.1523575113
Last articles
Cl in 5SYWCl in 5SYX
Cl in 5SYY
Cl in 5SYM
Cl in 5SYV
Cl in 5SYU
Cl in 5SYL
Cl in 5SYK
Cl in 5SYJ
Cl in 5SYI