Copper »
PDB 4e9v-4hhg »
4hd4 »
Copper in PDB 4hd4: Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant
Protein crystallography data
The structure of Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant, PDB code: 4hd4
was solved by
M.Goldfeder,
M.Kanteev,
N.Adir,
A.Fishman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.45 / 1.80 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 48.000, 78.580, 85.890, 90.00, 106.13, 90.00 |
| R / Rfree (%) | 20.7 / 22.8 |
Copper Binding Sites:
The binding sites of Copper atom in the Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant
(pdb code 4hd4). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant, PDB code: 4hd4:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant, PDB code: 4hd4:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 4hd4
Go back to
Copper binding site 1 out
of 2 in the Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant
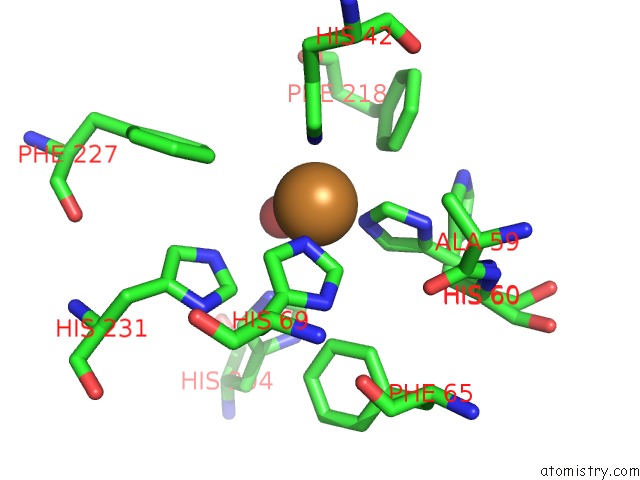
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant within 5.0Å range:
|
Copper binding site 2 out of 2 in 4hd4
Go back to
Copper binding site 2 out
of 2 in the Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant
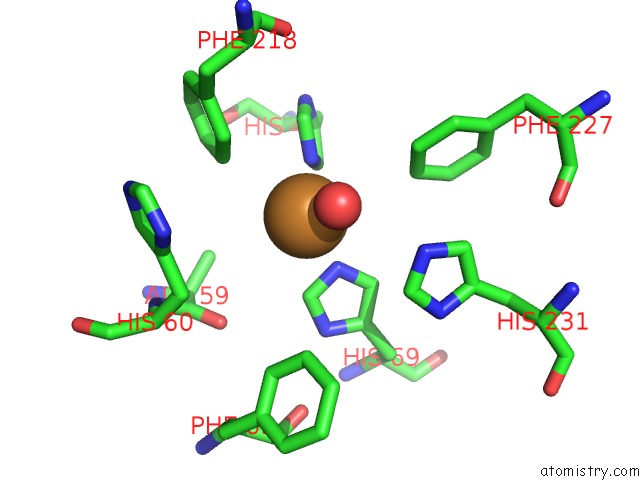
Mono view
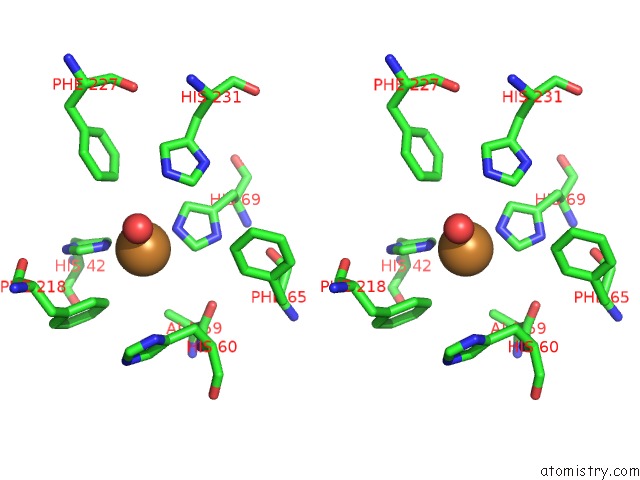
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Crystal Structure of Tyrosinase From Bacillus Megaterium V218F Mutant within 5.0Å range:
|
Reference:
M.Goldfeder,
M.Kanteev,
N.Adir,
A.Fishman.
Influencing the Monophenolase/Diphenolase Activity Ratio in Tyrosinase. Biochim.Biophys.Acta V.1834 629 2013.
ISSN: ISSN 0006-3002
PubMed: 23305929
DOI: 10.1016/J.BBAPAP.2012.12.021
Page generated: Mon Jul 14 03:44:47 2025
ISSN: ISSN 0006-3002
PubMed: 23305929
DOI: 10.1016/J.BBAPAP.2012.12.021
Last articles
Fe in 2WH8Fe in 2WGY
Fe in 2WB1
Fe in 2WCI
Fe in 2WD4
Fe in 2WAQ
Fe in 2WBP
Fe in 2W8S
Fe in 2WBO
Fe in 2WA4