Copper »
PDB 2vr7-2xv0 »
2xmt »
Copper in PDB 2xmt: Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form)
Protein crystallography data
The structure of Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form), PDB code: 2xmt
was solved by
A.Badarau,
S.J.Firbank,
A.A.Mccarthy,
M.J.Banfield,
C.Dennison,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.00 / 1.50 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 29.090, 29.660, 38.430, 100.89, 92.58, 117.91 |
| R / Rfree (%) | 16.041 / 20.279 |
Copper Binding Sites:
The binding sites of Copper atom in the Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form)
(pdb code 2xmt). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form), PDB code: 2xmt:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form), PDB code: 2xmt:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 2xmt
Go back to
Copper binding site 1 out
of 2 in the Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form)

Mono view
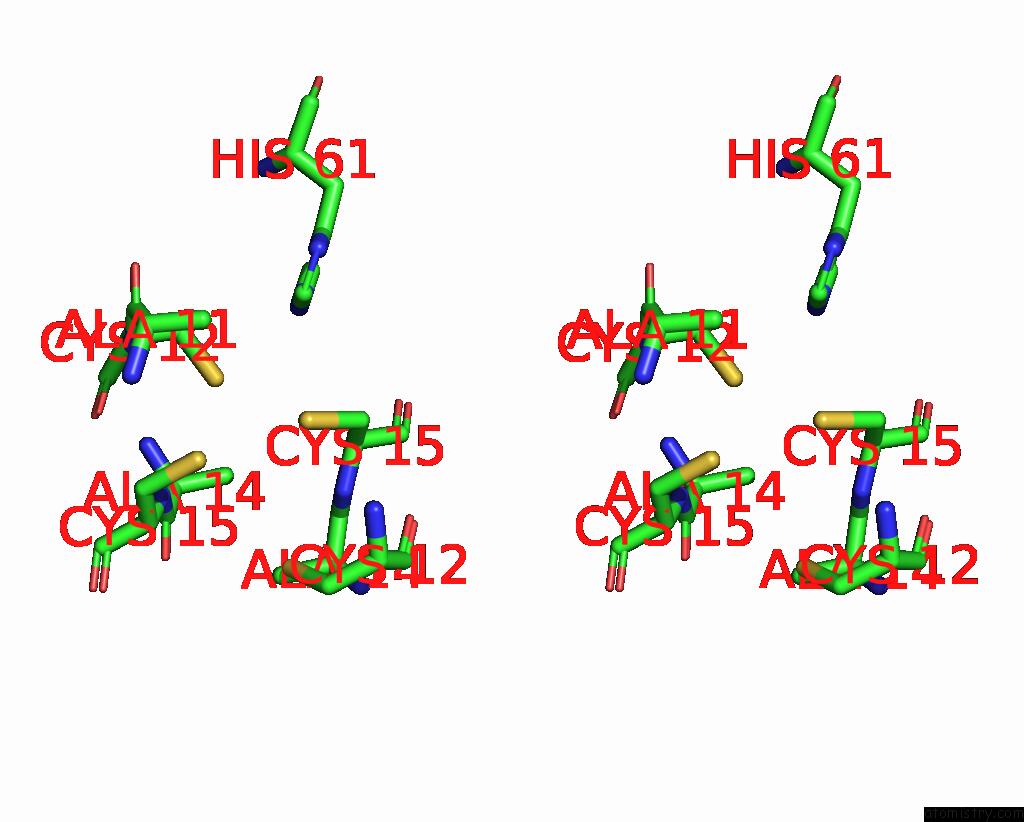
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form) within 5.0Å range:
|
Copper binding site 2 out of 2 in 2xmt
Go back to
Copper binding site 2 out
of 2 in the Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form)
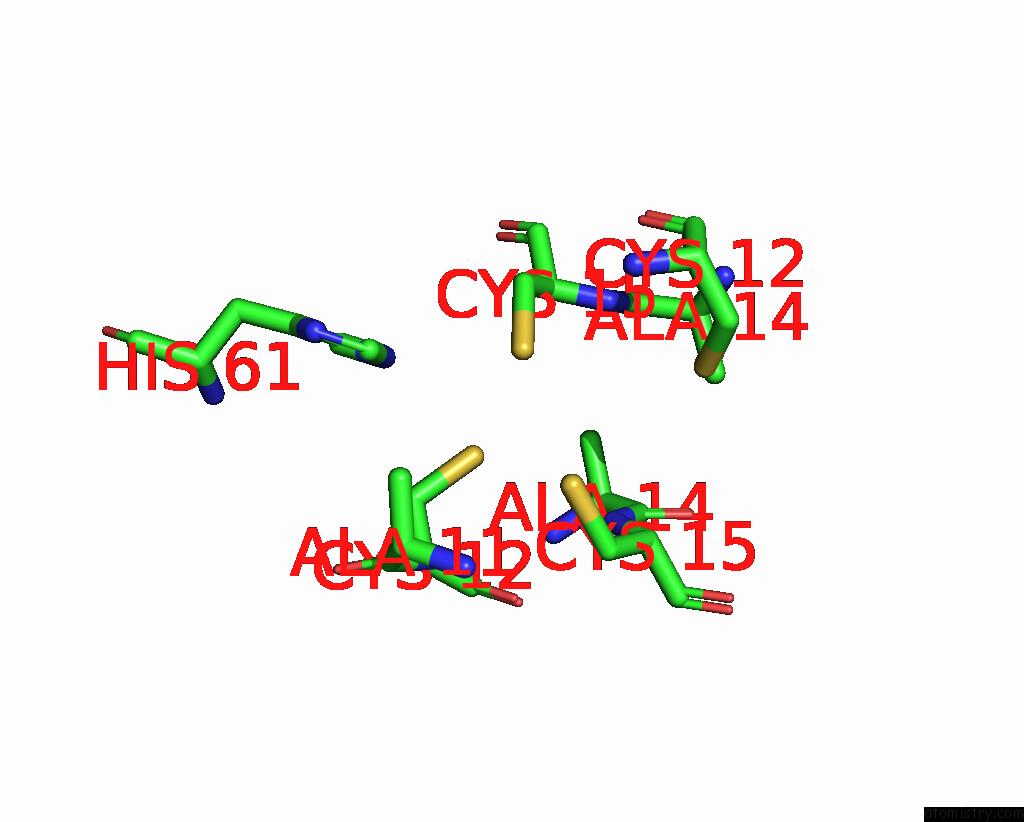
Mono view
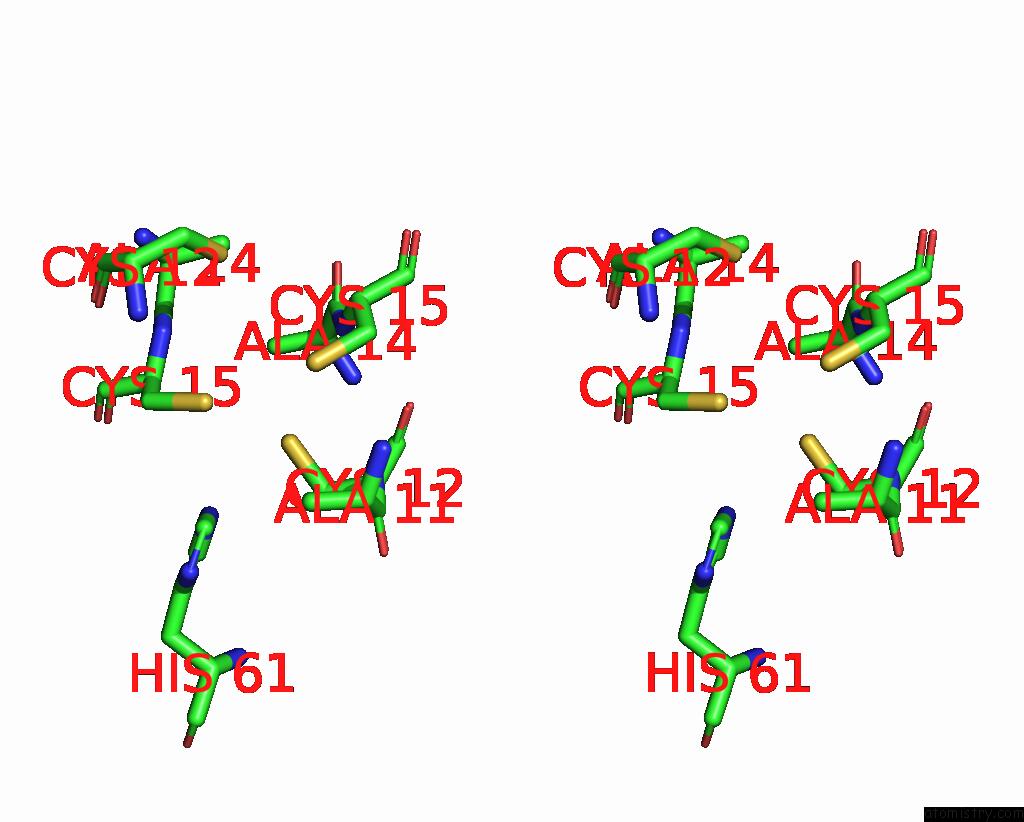
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of Copper Chaperone ATX1 From Synechocystis PCC6803 (CU1 Form) within 5.0Å range:
|
Reference:
A.Badarau,
S.J.Firbank,
A.A.Mccarthy,
M.J.Banfield,
C.Dennison.
Visualizing the Metal-Binding Versatility of Copper Trafficking Sites . Biochemistry V. 49 7798 2010.
ISSN: ISSN 0006-2960
PubMed: 20726513
DOI: 10.1021/BI101064W
Page generated: Wed Jul 31 00:11:49 2024
ISSN: ISSN 0006-2960
PubMed: 20726513
DOI: 10.1021/BI101064W
Last articles
Cl in 7WLWCl in 7WL9
Cl in 7WKZ
Cl in 7WKR
Cl in 7WGP
Cl in 7WK1
Cl in 7WHH
Cl in 7WGT
Cl in 7WF6
Cl in 7WGL