Copper »
PDB 2idq-2pp7 »
2lu5 »
Copper in PDB 2lu5: Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr)
Enzymatic activity of Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr)
All present enzymatic activity of Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr):
1.15.1.1;
1.15.1.1;
Copper Binding Sites:
The binding sites of Copper atom in the Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr)
(pdb code 2lu5). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr), PDB code: 2lu5:
In total only one binding site of Copper was determined in the Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr), PDB code: 2lu5:
Copper binding site 1 out of 1 in 2lu5
Go back to
Copper binding site 1 out
of 1 in the Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr)
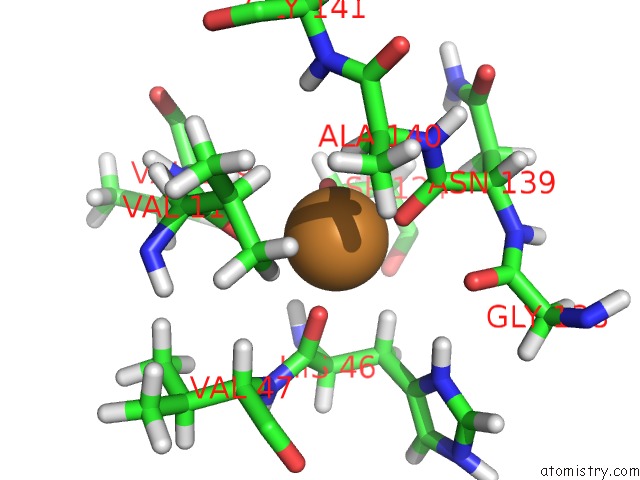
Mono view
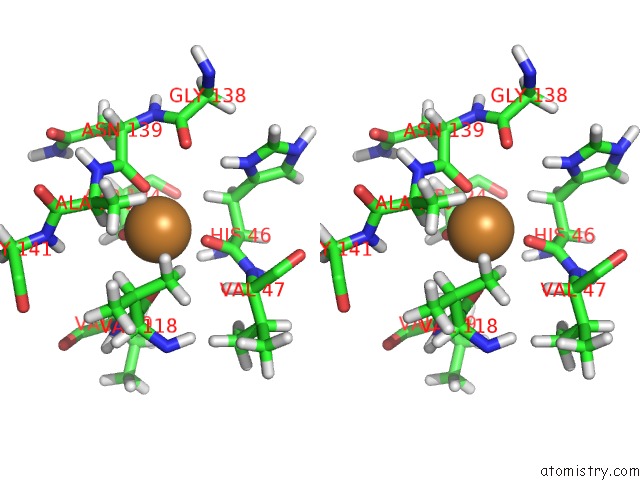
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Structure and Chemical Shifts of Cu(I),Zn(II) Superoxide Dismutase By Solid-State uc(Nmr) within 5.0Å range:
|
Reference:
M.J.Knight,
A.J.Pell,
I.Bertini,
I.C.Felli,
L.Gonnelli,
R.Pierattelli,
T.Herrmann,
L.Emsley,
G.Pintacuda.
Structure and Backbone Dynamics of A Microcrystalline Metalloprotein By Solid-State uc(Nmr). Proc.Natl.Acad.Sci.Usa V. 109 11095 2012.
ISSN: ISSN 0027-8424
PubMed: 22723345
DOI: 10.1073/PNAS.1204515109
Page generated: Tue Jul 30 23:50:23 2024
ISSN: ISSN 0027-8424
PubMed: 22723345
DOI: 10.1073/PNAS.1204515109
Last articles
Cl in 3DLECl in 3DKX
Cl in 3DL7
Cl in 3DL1
Cl in 3DKK
Cl in 3DKH
Cl in 3DKG
Cl in 3DKE
Cl in 3DKF
Cl in 3DKC