Copper »
PDB 2fqd-2idf »
2fwl »
Copper in PDB 2fwl: The Cytochrome C552/Cua Complex From Thermus Thermophilus
Enzymatic activity of The Cytochrome C552/Cua Complex From Thermus Thermophilus
All present enzymatic activity of The Cytochrome C552/Cua Complex From Thermus Thermophilus:
1.9.3.1;
1.9.3.1;
Other elements in 2fwl:
The structure of The Cytochrome C552/Cua Complex From Thermus Thermophilus also contains other interesting chemical elements:
| Iron | (Fe) | 3 atoms |
Copper Binding Sites:
The binding sites of Copper atom in the The Cytochrome C552/Cua Complex From Thermus Thermophilus
(pdb code 2fwl). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total 2 binding sites of Copper where determined in the The Cytochrome C552/Cua Complex From Thermus Thermophilus, PDB code: 2fwl:
Jump to Copper binding site number: 1; 2;
In total 2 binding sites of Copper where determined in the The Cytochrome C552/Cua Complex From Thermus Thermophilus, PDB code: 2fwl:
Jump to Copper binding site number: 1; 2;
Copper binding site 1 out of 2 in 2fwl
Go back to
Copper binding site 1 out
of 2 in the The Cytochrome C552/Cua Complex From Thermus Thermophilus
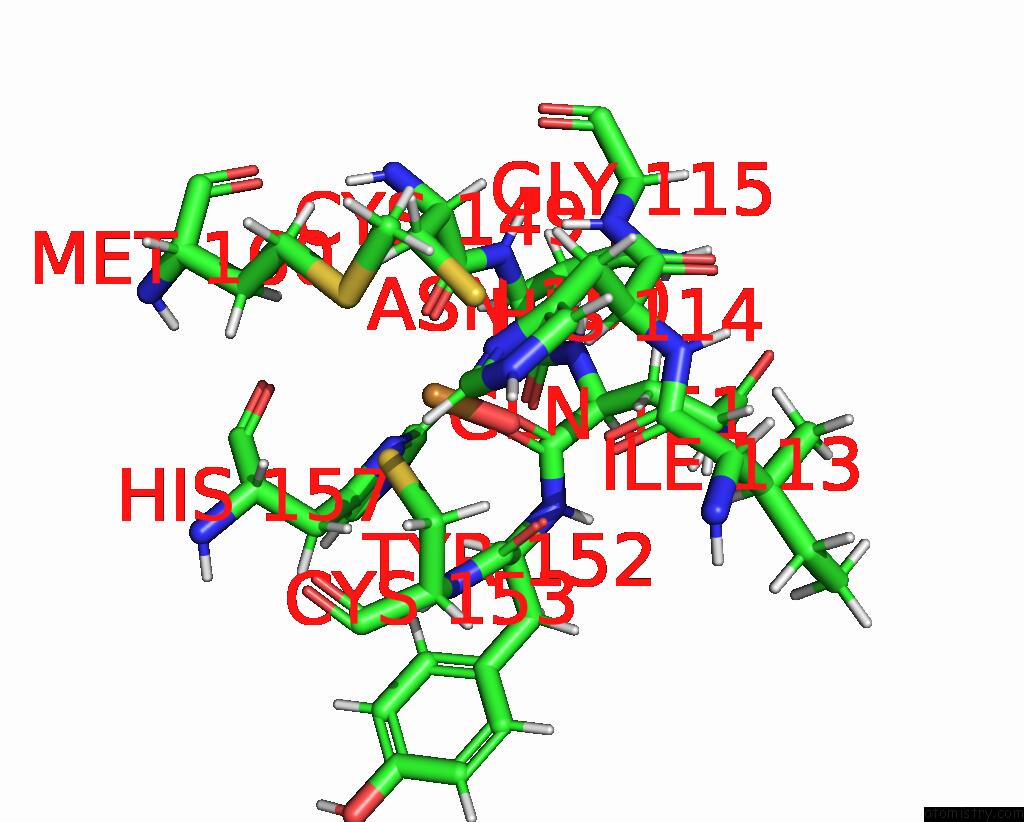
Mono view
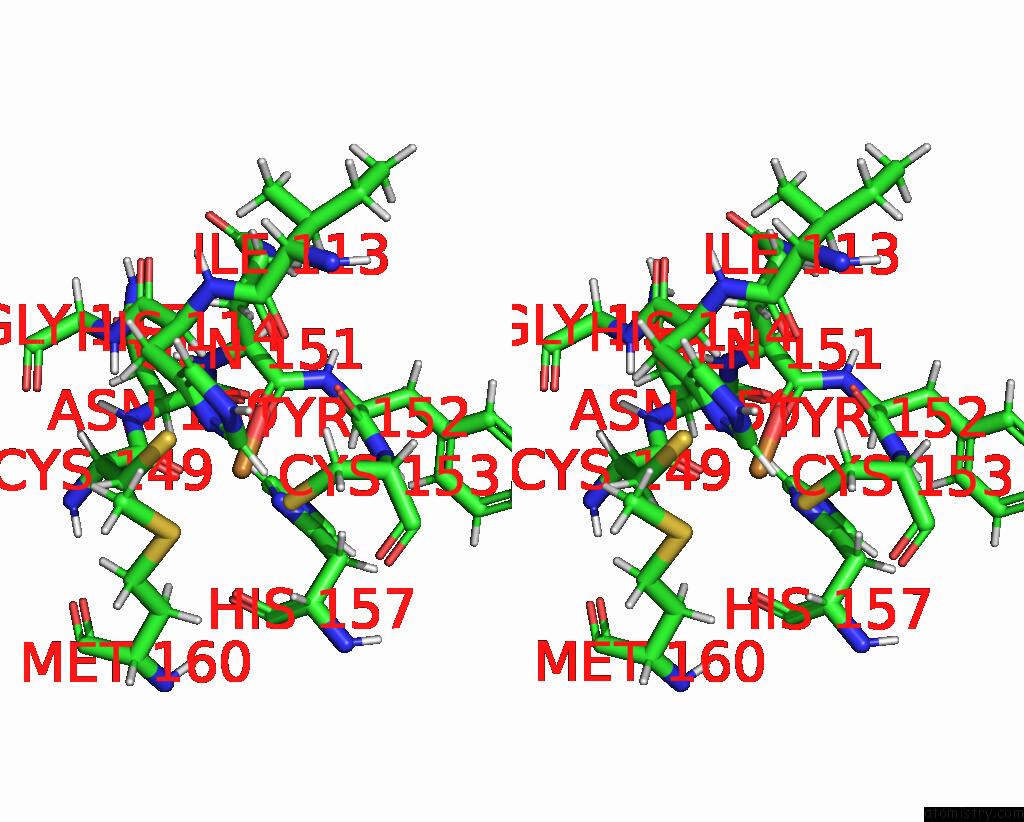
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of The Cytochrome C552/Cua Complex From Thermus Thermophilus within 5.0Å range:
|
Copper binding site 2 out of 2 in 2fwl
Go back to
Copper binding site 2 out
of 2 in the The Cytochrome C552/Cua Complex From Thermus Thermophilus
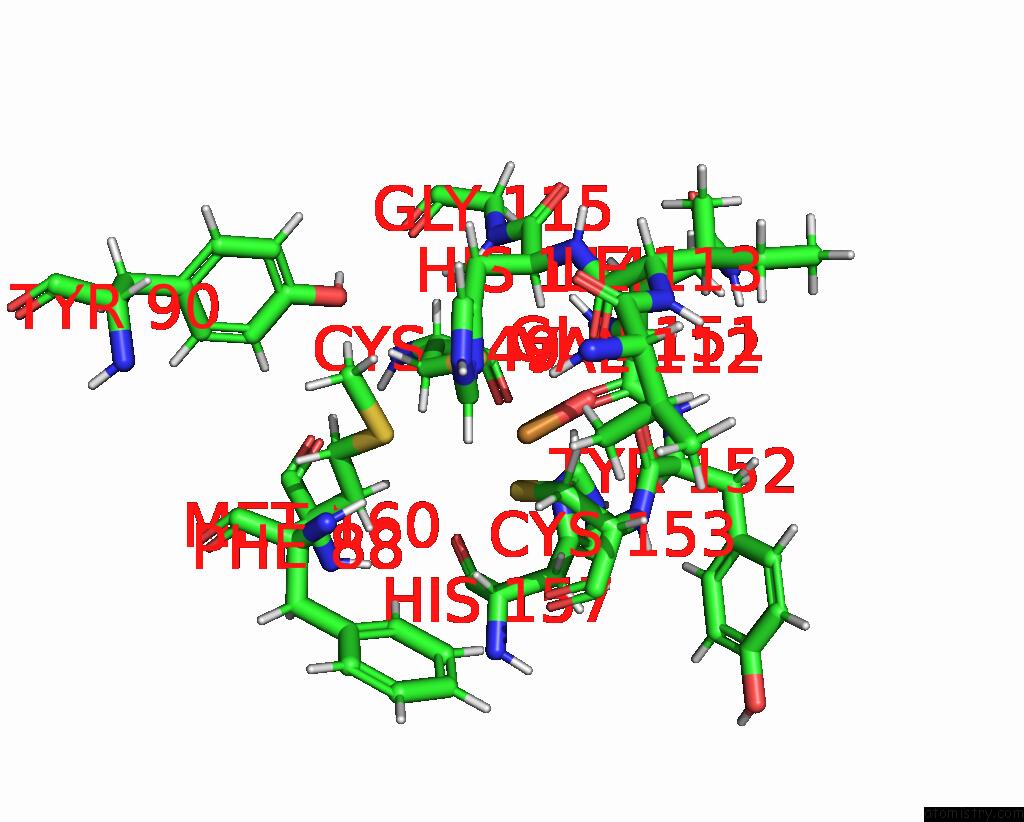
Mono view
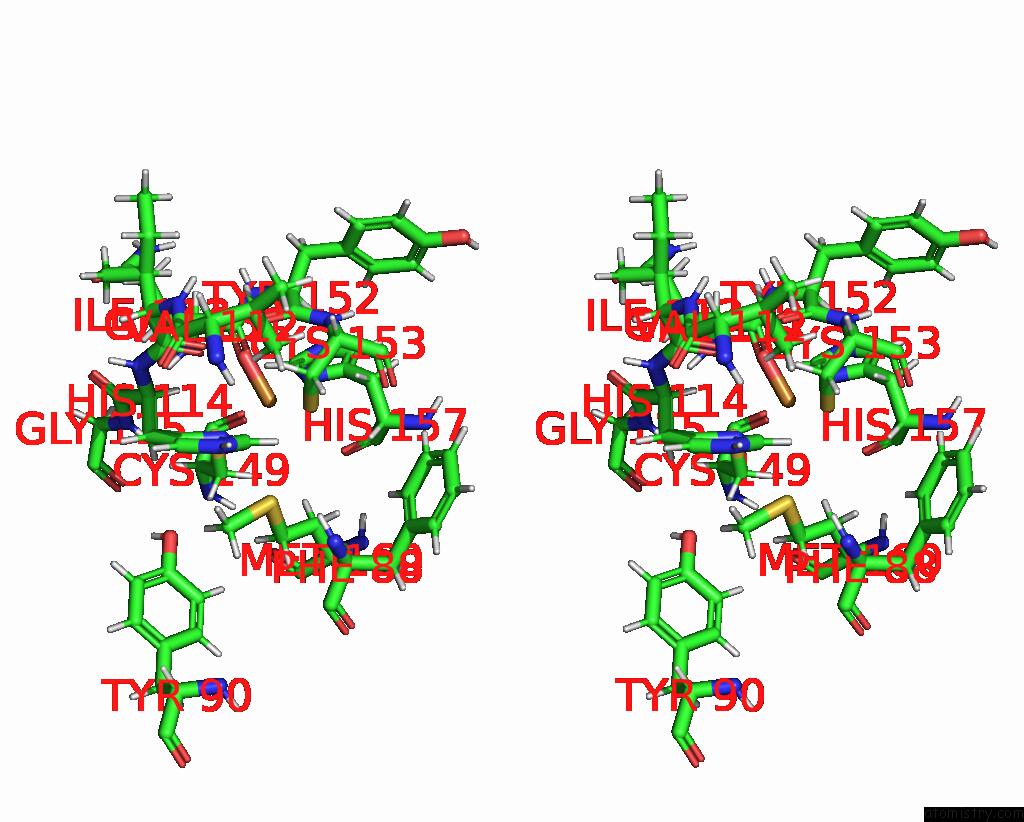
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 2 of The Cytochrome C552/Cua Complex From Thermus Thermophilus within 5.0Å range:
|
Reference:
L.Muresanu,
P.Pristovsek,
F.Loehr,
O.Maneg,
M.D.Mukrasch,
H.Rueterjans,
B.Ludwig,
C.Luecke.
The Electron Transfer Complex Between Cytochrome C552 and the Cua Domain of the Thermus Thermophilus BA3 Oxidase - A Combined uc(Nmr) and Computational Approach J.Biol.Chem. V. 281 14503 2006.
ISSN: ISSN 0021-9258
PubMed: 16554303
DOI: 10.1074/JBC.M601108200
Page generated: Tue Jul 30 23:34:39 2024
ISSN: ISSN 0021-9258
PubMed: 16554303
DOI: 10.1074/JBC.M601108200
Last articles
Ca in 5VCOCa in 5VCN
Ca in 5VC1
Ca in 5VA9
Ca in 5VBJ
Ca in 5VAO
Ca in 5V8K
Ca in 5V6V
Ca in 5V2C
Ca in 5V4D