Copper »
PDB 1x9r-2ahl »
1zm5 »
Copper in PDB 1zm5: Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure
Protein crystallography data
The structure of Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure, PDB code: 1zm5
was solved by
R.Boer,
S.Russi,
A.Guasch,
M.Lucas,
A.G.Blanco,
M.Coll,
F.De Lacruz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.68 / 2.50 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.077, 91.077, 205.551, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 23.3 / 27.6 |
Copper Binding Sites:
The binding sites of Copper atom in the Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure
(pdb code 1zm5). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure, PDB code: 1zm5:
In total only one binding site of Copper was determined in the Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure, PDB code: 1zm5:
Copper binding site 1 out of 1 in 1zm5
Go back to
Copper binding site 1 out
of 1 in the Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure
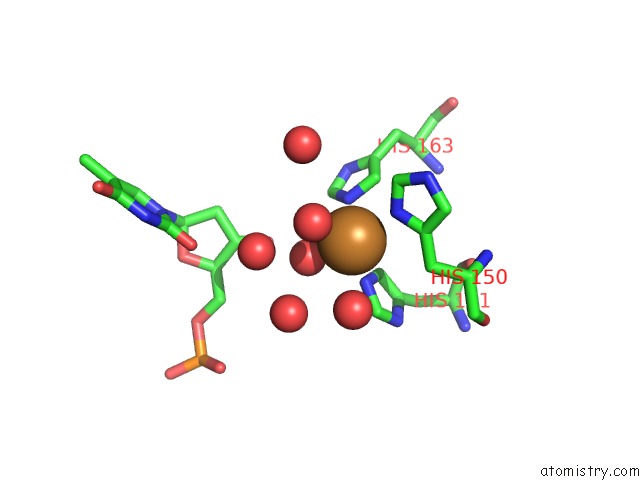
Mono view
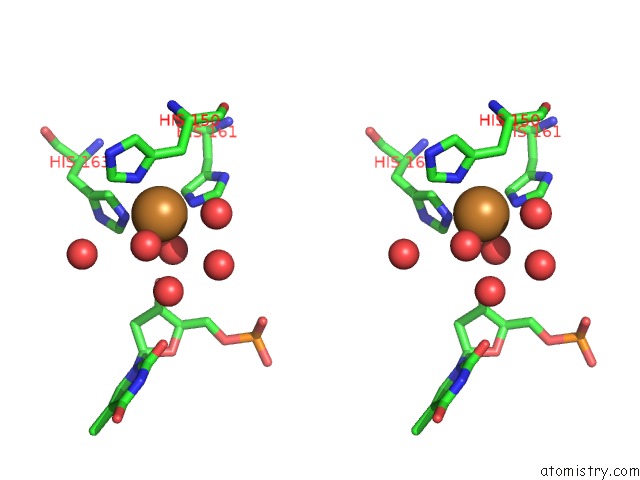
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Conjugative Relaxase Trwc in Complex with Orit Dna, Cooper- Bound Structure within 5.0Å range:
|
Reference:
R.Boer,
S.Russi,
A.Guasch,
M.Lucas,
A.G.Blanco,
R.Perez-Luque,
M.Coll,
F.De La Cruz.
Unveiling the Molecular Mechanism of A Conjugative Relaxase: the Structure of Trwc Complexed with A 27-Mer Dna Comprising the Recognition Hairpin and the Cleavage Site J.Mol.Biol. V. 358 857 2006.
ISSN: ISSN 0022-2836
PubMed: 16540117
DOI: 10.1016/J.JMB.2006.02.018
Page generated: Tue Jul 30 23:05:28 2024
ISSN: ISSN 0022-2836
PubMed: 16540117
DOI: 10.1016/J.JMB.2006.02.018
Last articles
Cl in 3F0ACl in 3F03
Cl in 3EYY
Cl in 3EZJ
Cl in 3EXN
Cl in 3EXI
Cl in 3EW5
Cl in 3EXC
Cl in 3EX6
Cl in 3EWK