Copper »
PDB 1tmx-1x9l »
1u96 »
Copper in PDB 1u96: Solution Structure of Yeast COX17 with Copper Bound
Copper Binding Sites:
The binding sites of Copper atom in the Solution Structure of Yeast COX17 with Copper Bound
(pdb code 1u96). This binding sites where shown within
5.0 Angstroms radius around Copper atom.
In total only one binding site of Copper was determined in the Solution Structure of Yeast COX17 with Copper Bound, PDB code: 1u96:
In total only one binding site of Copper was determined in the Solution Structure of Yeast COX17 with Copper Bound, PDB code: 1u96:
Copper binding site 1 out of 1 in 1u96
Go back to
Copper binding site 1 out
of 1 in the Solution Structure of Yeast COX17 with Copper Bound
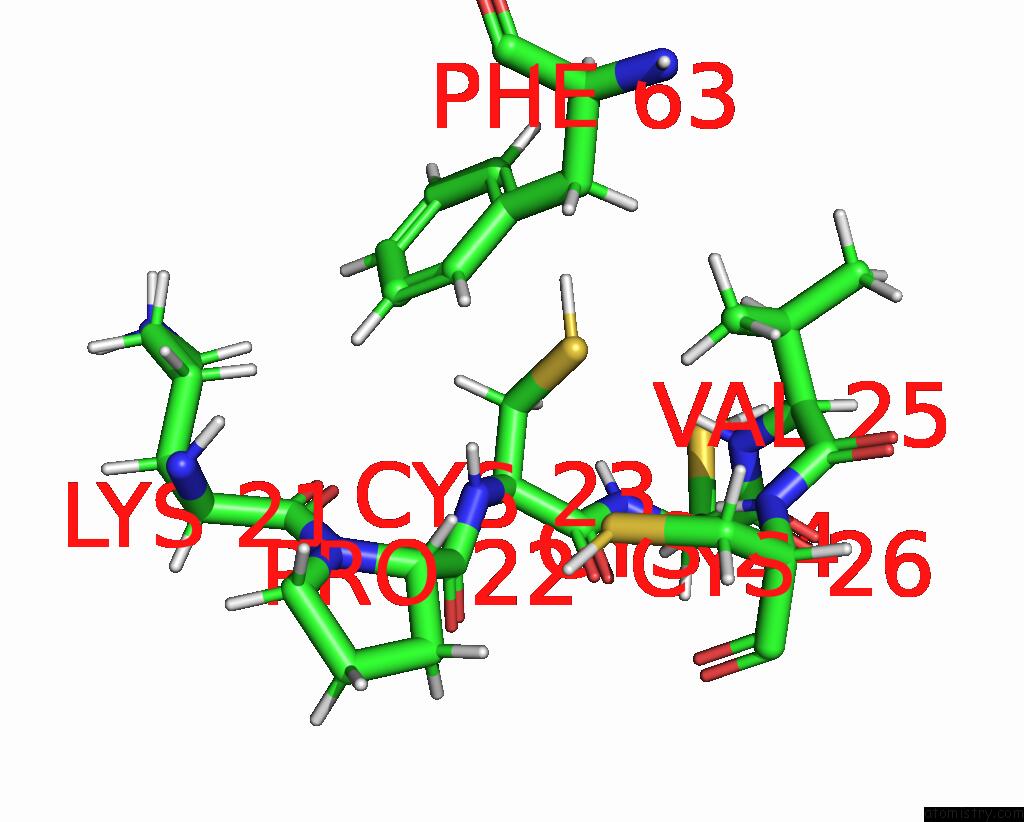
Mono view
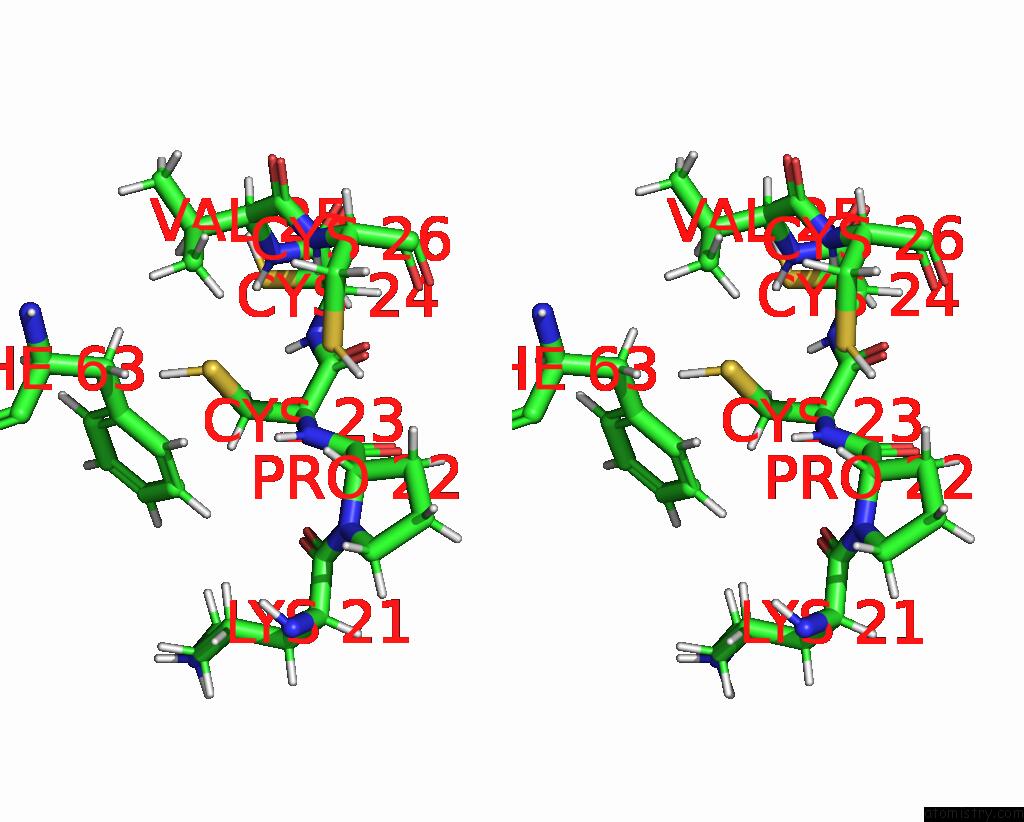
Stereo pair view

Mono view

Stereo pair view
A full contact list of Copper with other atoms in the Cu binding
site number 1 of Solution Structure of Yeast COX17 with Copper Bound within 5.0Å range:
|
Reference:
C.Abajian,
L.A.Yatsunyk,
B.E.Ramirez,
A.C.Rosenzweig.
Yeast COX17 Solution Structure and Copper(I) Binding. J.Biol.Chem. V. 279 53584 2004.
ISSN: ISSN 0021-9258
PubMed: 15465825
DOI: 10.1074/JBC.M408099200
Page generated: Tue Jul 30 22:51:38 2024
ISSN: ISSN 0021-9258
PubMed: 15465825
DOI: 10.1074/JBC.M408099200
Last articles
Cl in 5RAYCl in 5RAX
Cl in 5RAW
Cl in 5RAV
Cl in 5RAU
Cl in 5RAS
Cl in 5RAR
Cl in 5RAQ
Cl in 5RAP
Cl in 5RAO